Notebook
Construction for Lysisbox
Tuesday, July 20 By: Wataru, Tomo, Yuki, Kazuya, Ken, Makoto
| Name | Well | Sample | Competent Cells | Total | Plate | Incubation | Colony
|
| J23100 | 1-18-C | 1 µL | 20 | 21 | LB (Amp+) | At 37℃, 7/20 20:50 - 7/21 17:00 | ○
|
| J23105 | 1-18-M | 1 | 20 | 21 | ○
|
| J23116 | 1-20-M | 1 | 20 | 21 | ○
|
| R0011 | 1-6-G | 1 | 20 | 21 | ○
|
| E0840 | 1-12-O | 1 | 20 | 21 | ○
|
| J06702 | 2-8-E | 1 | 20 | 21 | ○
|
| pSB4K5 | 1-5-G | 1 | 20 | 21 | ×
|
| B0015 | 1-23-L | 1 | 20 | 21 | LB (Kan+) | ×
|
A vector of pSB4K5 is Kanamycin-resistance, however, we plated it to LB plate (Amp+). And We didn't pre-culture B0015 despite its vector is Kanamycin-resistance. So, it was predicted that we will fail the transformation of pSB4K5 and B0015.
Wednesday, July 21 By: Wataru, Ken, Makoto, Takuya Y.
Culture at 37℃ from 07/21 20:50 to 07/22 17:00 and Making Master Plate
| Name | Well | Sample | Competent Cells | Total | Plate | Incubation | Colony
|
| pSB4K5 | 1-5-G | 1 µL | 20 | 21 | LB (Kan+) | At 37℃, 7/21 20:50 - 7/22 16:30 | ○
|
| B0015 | 1-23-L | 1 | 20 | 21 | ○
|
PCR for SRRz and S
| No. | Water | MgSO4 | dNTPs | 10xBuffer | Template DNA | Primer Fwd. | Primer Rev. (SRRz) | Primer Rev. (S) | KOD Plus ver.2 | Total
|
| 1 | 28 µL | 3 | 5 | 5 | 5 | 1.5 | 1.5 | - | 1 | 50
|
| 2 | 28 | 3 | 5 | 5 | 5 | 1.5 | 1.5 | - | 1 | 50
|
| 3 | 28 | 3 | 5 | 5 | 5 | 1.5 | - | 1.5 | 1 | 50
|
| 4 | 28 | 3 | 5 | 5 | 5 | 1.5 | - | 1.5 | 1 | 50
|
| 5 | 28 | 3 | 5 | 5 | 5 | 1.5 | 1.5 | - | 1 | 50
|
| 6 | 28 | 3 | 5 | 5 | 5 | 1.5 | 1.5 | - | 1 | 50
|
| 7 | 28 | 3 | 5 | 5 | 5 | 1.5 | - | 1.5 | 1 | 50
|
| 8 | 28 | 3 | 5 | 5 | 5 | 1.5 | - | 1.5 | 1 | 50
|
| 94℃ | 2min |
|
| 98℃ | 10s | 30 cycles
|
| 55℃ | 30s
|
| 68℃ | 4min
|
| 4℃ | forever |
|
Thursday, July 22 By: Wataru
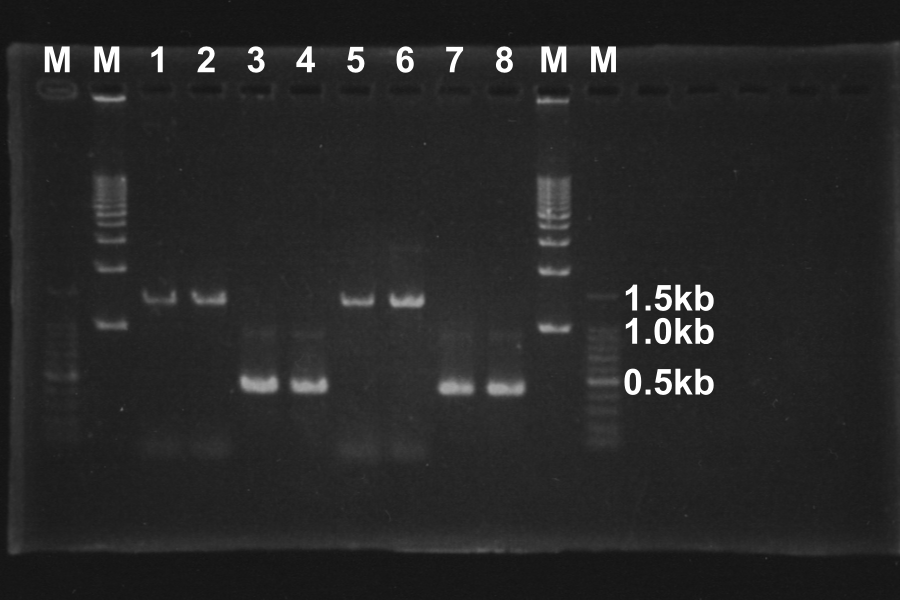
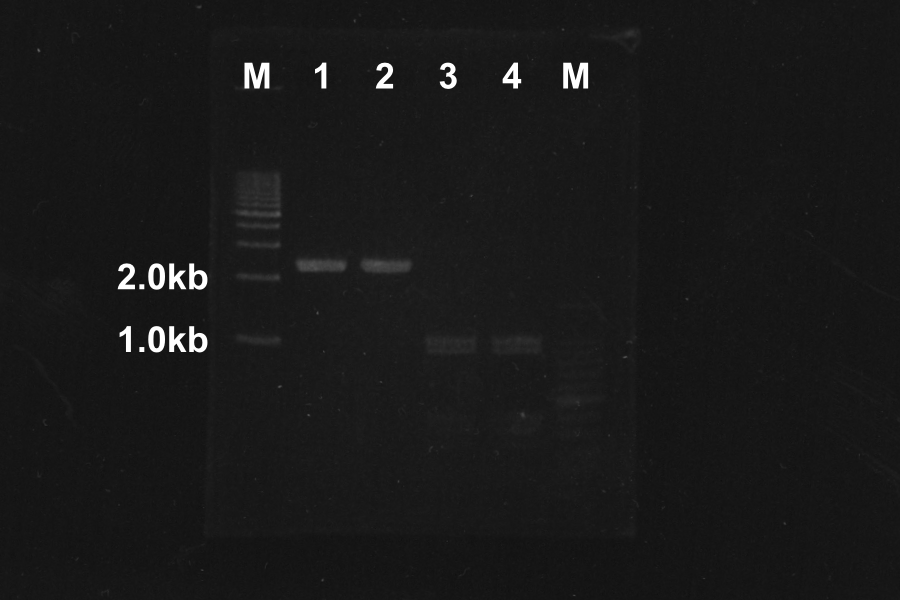
| No. | Name | Length(bp) | Result
|
| 1 | SRRz | 1386 | ○
|
| 2 | SRRz | 1386 | ○
|
| 3 | S | 442 | ○
|
| 4 | S | 442 | ○
|
| 5 | SRRz | 1386 | ○
|
| 6 | SRRz | 1386 | ○
|
| 7 | S | 442 | ○
|
| 8 | S | 442 | ○
|
Marker: 100bp, 1kb, 1kb, 100bp.
| Name | Concentration
|
| J23100 | 18.5 (ng/µL)
|
| J23105 | 12.5
|
| J23116 | 14.6
|
| R0011 | 8.6
|
| E0840 | 12.1
|
| J06702 | 14.7
|
The concentration of all samples was very week. Probably our shaking incubation was week.
Culture from 07/22 17:00 to 07/23 10:00 and Making Master Plates of pSB4K5 and B0015
Friday, July 23 By: Wataru, Tomo, Makoto
| Name | Concentration
|
| pSB4K5 | 79.2 (ng/µL)
|
| B0015 | -
|
We lost B0015 by our mistake. The concentration of pSB4K5 is high, so this condition of shaking incubation is moderate.
| No. | Name | Concentration | New Name
|
| 1 | SRRz | 18.6 ng/µL | -
|
| 3 | S | 77.6 | SSam7(1)
|
| 5 | SRRz | 33.6 | -
|
| 7 | S | 65.4 | SSam7(2)
|
The concentration of sample number 1 and 5, the PCR products of S-R-Rz/Rz1, is week, so we desided to retry PCR.
PCR for SRRz
| No. | Water | MgSO4 | dNTPs | 10xBuffer | Template DNA | Primer Fwd. (SRRz) | Primer Rev. (SRRz) | KOD plus ver.2 | Total
|
| 1 | 28 µL | 3 | 5 | 5 | 5 | 1.5 | 1.5 | 1 | 50
|
| 2 | 28 | 3 | 5 | 5 | 5 | 1.5 | 1.5 | 1 | 50
|
| 3 | 26.5 | 4.5 | 5 | 5 | 5 | 1.5 | 1.5 | 1 | 50
|
| 4 | 26.5 | 4.5 | 5 | 5 | 5 | 1.5 | 1.5 | 1 | 50
|
| 5 | 25 | 6 | 5 | 5 | 5 | 1.5 | 1.5 | 1 | 50
|
| 6 | 25 | 6 | 5 | 5 | 5 | 1.5 | 1.5 | 1 | 50
|
| 94℃ | 2min |
|
| 98℃ | 10s | 30 cycles
|
| 55℃ | 30s
|
| 68℃ | 4min
|
| 4℃ | forever |
|
| No. | Name | Sample | 10xBuffer | BSA | Enzyme | MilliQ | Total | Incubation
|
| 1 | J06702 | 5 µL | 1 | 0.1 | EcoRI | 0.1 | 3.6 | 10 | At 37℃ 7/23 18:00 - 7/23 18:30
|
| 2 | J06702 | 5 | 1 | 0.1 | XbaI | 0.1 | 3.6 | 10
|
| 3 | J06702 | 5 | 1 | 0.1 | SpeI | 0.1 | 3.6 | 10
|
| 4 | J06702 | 5 | 1 | 0.1 | PstI | 0.1 | 3.6 | 10
|
| 5 | J06702 | 5 | 1 | 0.1 | - | 3.7 | 10
|
Marker: 1kb.
Comparison to No. 5 (control, circular DNA), the bands of No. 1, 2, 3, and 4 was shifted. The DNA of them was linearized by Restriction enzymes. So, our restriction enzymes work correctly.
| Name | Sample | 10xBuffer | Enzyme 1 | Enzyme 2 | MilliQ | Total | Incubation
|
| SSam7(1) | 11 µL | 5 | EcoRI | 0.2 | SpeI | 0.2 | 33.6 | 50 | At 37℃ for 2h
|
| SSam7(2) | 11 | 5 | EcoRI | 0.2 | SpeI | 0.2 | 33.6 | 50
|
| E0840 | 45 | 5 | EcoRI | 0.2 | XbaI | 0.2 | 0 | 50
|
After PCR Purification, evaporated them and diluted 3µL.
| Name | Vector | Insert | Ligation High | Total
|
| SSam7(1)-E0840 | E0840 | 0.5µL | SSam7(1) | 0.5 | 1 | 2
|
| SSam7(2)-E0840 | E0840 | 0.5 | SSam7(2) | 0.5 | 1 | 2
|
Monday, July 26 By: Wataru, Tomonori, Makoto
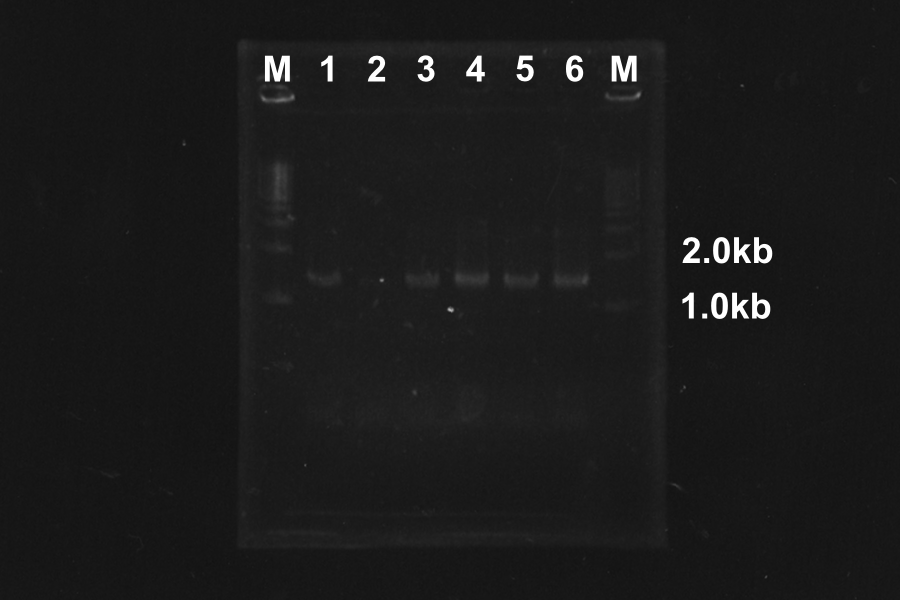
| No. | Name | Length(bp) | Result
|
| 1 | SRRz | 1386 |
|
| 2 | SRRz | 1386 |
|
| 3 | SRRz | 1386 |
|
| 4 | SRRz | 1386 |
|
| 5 | SRRz | 1386 |
|
| 6 | SRRz | 1386 |
|
Marker: 1kb.
At the condition 4 (4.5µL MgSO4) and 6 (6µL MgSO4), SRRz is amplified very much. So we decided to use them.
PCR Purification
| No. | Name | Concentration | New Name
|
| 4 | SRRZ | 51.6 ng/µL | SRRzSam7(1)
|
| 5 | SRRZ | 59.3 |
|
| 6 | SRRZ | 59.6 | SRRzSam7(2)
|
| Name | Well | Sample | Competent Cell | Total | Plate | Incubation | Colony
|
| E0240 | 1-12-M | 1 µL | 20 | 21 | LB (Amp+) | At 37℃ 7/26 - 7/27 | ×
|
| I20260 | 2-17-F | 1 | 20 | 21 | LB (Kan+) | ×
|
| J04450 | 1-5-E | 1 | 20 | 21 | ×
|
Culture of pSB4K5, E0840, and B0015
Tuesday, July 27 By: Wataru, Tomo, Kazuya, Ken, Naoi
Colony PCR of SSam7-E0840 (Electrophoresis for 35min)
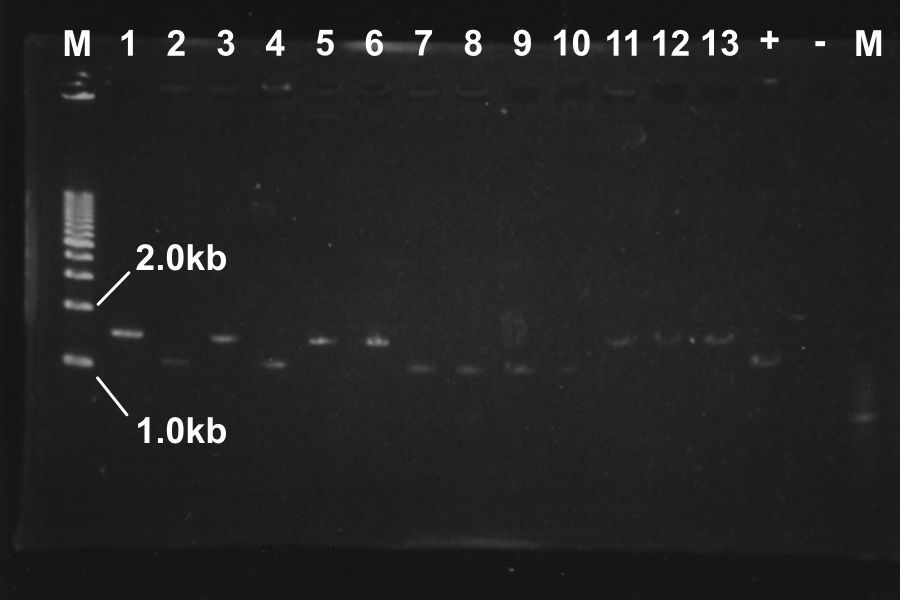
| No. | Name | Length | Result
|
| 1 | SSam7(1)-E0840 | 1522 | ○
|
| 2 | SSam7(1)-E0840 | 1522 | ×
|
| 3 | SSam7(1)-E0840 | 1522 | ○
|
| 4 | SSam7(1)-E0840 | 1522 | ×
|
| 5 | SSam7(1)-E0840 | 1522 | ○
|
| 6 | SSam7(1)-E0840 | 1522 | ◎ (Use as SSam7(1)-E0840)
|
| 7 | SSam7(2)-E0840 | 1522 | ×
|
| 8 | SSam7(2)-E0840 | 1522 | ×
|
| 9 | SSam7(2)-E0840 | 1522 | ×
|
| 10 | SSam7(2)-E0840 | 1522 | ×
|
| 11 | SSam7(2)-E0840 | 1522 | ◎ (Use as SSam7(2)-E0840)
|
| 12 | SSam7(2)-E0840 | 1522 | ○
|
| 13 | SSam7(2)-E0840 | 1522 | ○
|
| + | E0840 | 1116 |
|
| - | None | |
|
Marker: 1kb, 100bp
| Name | Concentration
|
| R0011 | 26.9 ng/µL
|
| B0015 | 120.0
|
| E0840 | 120.1
|
| Name | Sample | 2 buffer | BSA | Enzyme 1 | Enzyme 2 | MilliQ | Total | Incubation
|
| B0015 | 30 µL | 5 | 0.5 | EcoRI | 0.4 | XbaI | 0.3 | 13.7 | 50 | At 37℃ 16:45 - 18:00
|
| SRRzSam7(1) | 40 | 5 | 0.5 | EcoRI | 0.4 | SpeI | 0.4 | 3.8 | 50
|
| SRRzSam7(2) | 40 | 5 | 0.5 | EcoRI | 0.4 | SpeI | 0.4 | 3.8 | 50
|
| Name | Sample | Competent Cells | Total | Plate | Incubation | Colony
|
| SRRzSam7(1)-B0015 | | | | | | ○
|
| SRRzSam7(2)-B0015 | | | | ○
|
Wednesday, July 28 By:
| Name | Concentration
|
| SSam7(1)-E0840 | 95.5 ng/µL
|
| SSam7(2)-E0840 | 98.6
|
Diluted SSam7(1)-E0840 and SSam7(2)-E0840 20 times with water, and used as template DNA.
Deletion PCR to delete a functional domain of S gene
| Water | MgSO4 | dNTPs | 10xBuffer | Primer Fwd. | Primer Rev. | Template (1) | Template (2) | KOD Plus ver.2 | Total
|
| SSam7,ΔTMD1(1)-E0840 (1) | 28 µL | 3 | 5 | 5 | 1.5 | 1.5 | 5 | - | 1 | 50
|
| SSam7,ΔTMD1(1)-E0840 (2) | 28 | 3 | 5 | 5 | 1.5 | 1.5 | 5 | - | 1 | 50
|
| SSam7,ΔTMD1(2)-E0840 (1) | 28 | 3 | 5 | 5 | 1.5 | 1.5 | - | 5 | 1 | 50
|
| SSam7,ΔTMD1(2)-E0840 (2) | 28 | 3 | 5 | 5 | 1.5 | 1.5 | - | 5 | 1 | 50
|
| 94℃ | 2min |
|
| 98℃ | 10s | 35 cycles
|
| 55℃ | 30s
|
| 68℃ | 4min
|
| 4℃ | forever |
|
| Name | Sample | fast digestion buffer | DpnI | MilliQ | Total
|
| SSam7(1)-E0840 | 3 µL | 1 | 0.1 | 5.8 | 10
|
| SSam7(2)-E0840 | 3 | 1 | 0.1 | 5.8 | 10
|
| No. | Name | Length | Result
|
| 1 | Not digested SSam7(1)-E0840 | 3363bp |
|
| 2 | Not digested SSam7(2)-E0840 | 3363 |
|
| 3 | Digested SSam7(1)-E0840 | 1021, 933, 402, 341, 258, 105, ... |
|
| 4 | Digested SSam7(2)-E0840 | 1021, 933, 402, 341, 258, 105, ... |
|
Marker: 1kb, 100bp
DpnI works correctly.
Thursday, July 29 By:
| Name | Sample volume | Fastdigestion Buffer | Enzyme 1 | MilliQ | Total | Incubation
|
| SSam7,ΔTMD1(1)-E0840 (1) | 50 µL | 6 | DpnI | 0.2 | 3.8 | 60 | 07/29 09:40 - 07/29 11:00
|
| SSam7,ΔTMD1(2)-E0840 (1) | 50 | 6 | DpnI | 0.2 | 3.8 | 60
|
| Name | Sample | MilliQ | Ligation High | T4 Kinase | Total | Incubation
|
| SSam7,ΔTMD1(1)-E0840 (1) | 2 µL | 7 | 5 | 1 | 15 | 07/29 11:30 ~ 07/29 13:00
|
| SSam7,ΔTMD1(2)-E0840 (1) | 2 | 7 | 5 | 1 | 15
|
| Name | Sample Volume | Competent Cell | Total | Plate | Incubation | Colony
|
| SSam7,ΔTMD1(1)-E0840 (1) | 3 µL | 30 | 33 | LB (Amp+) | 07/29 ~ 07/30 | ○
|
| SSam7,ΔTMD1(2)-E0840 (1) | 3 | 30 | 33 | ○
|
Monday, August 2 By: Wataru, Ken
| Name | Concentration
|
| SSam7,ΔTMD1-E0840 (1) | 52.7 ng/µL
|
| SSam7,ΔTMD1-E0840 (2) | 54.4
|
| SSam7,ΔTMD1-E0840 (3) | 89.5
|
| pSB4K5 | 50.7
|
| R0011 | 18.6
|
E0240 is very important parts to measure RPU of promoters in iGEM. However, we failed to transfect it to E.coli from parts kit of iGEM. So we decided to amplify this parts by PCR.
| Name | Water | MgSO4 | dNTPs | 10xBuffer | Primer VF2 | Primer VR | Template E0240 | KOD Pllus ver.2 | Total
|
| E0240(1) | 28 µL | 3 | 5 | 5 | 1.5 | 1.5 | 5 | 1 | 50
|
| E0240(2) | 28 | 3 | 5 | 5 | 1.5 | 1.5 | 5 | 1 | 50
|
| 94℃ | 2min |
|
| 98℃ | 10s | 35 cycles
|
| 55℃ | 30s
|
| 68℃ | 4min
|
| 4℃ | forever |
|
| Name | Concentration
|
| E0240(1) | 42.6 ng/µL
|
| E0240(2) | 55.3
|
Restriction Digestion for inserting E0240 to pSB4K5 by 3A assembly
| Name | Sample volume | 2 buffer | BSA | Enzyme 1 | Enzyme 2 | MilliQ | Total
|
| E0240(1) [XP] | 30 µL | 5 | 0.5 | XbaI | 0.2 | PstI | 0.2 | 14.1 | 50
|
| E0240(2) [XP] | 30 | 5 | 0.5 | XbaI | 0.2 | PstI | 0.2 | 14.1 | 50
|
PCR Purification
| Name | Concentration | Volume
|
| E0240(1) [XP] | 21.8 ng/µL | 40 µL
|
| E0240(2) [XP] | 32.4 | 45
|
Stored at -20℃.
Error PCR
| Name | Water | MgSO4 | dNTPs | 10xBuffer | Primer VF2 | Primer VR | Template (1) | Template (2) | Template (3) | KOD Pllus ver.2 | Total
|
| SSam7,ΔTMD1-E0840 (1) | 32 µL | 3 | 5 | 5 | 1.5 | 1.5 | 1 | - | - | 1 | 50
|
| SSam7,ΔTMD1-E0840 (2) | 32 | 3 | 5 | 5 | 1.5 | 1.5 | - | 1 | - | 1 | 50
|
| SSam7,ΔTMD1-E0840 (3) | 32 | 3 | 5 | 5 | 1.5 | 1.5 | - | - | 1 | 1 | 50
|
| 94℃ | 2min |
|
| 98℃ | 10s | 20 cycles
|
| 68℃ | 4min
|
| 4℃ | forever |
|
| Name | Sample | Competent Cells | Total | Plate | Incubation | Colony
|
| SSam7,ΔTMD1-E0840 (1) | 2 µL | 20 | 22 | - | - | ○
|
| SSam7,ΔTMD1-E0840 (2) | 2 | 20 | 22 | }
|
| SSam7,ΔTMD1-E0840 (3) | 2 | 20 | 22 | ○
|
Tuesday, August 3 By:
Culture
Picked two colonies from SSam7,ΔTMD1-E0840 (1), and SSam7,ΔTMD1-E0840 (3), and cultured at 37℃ from 08/03 to 08/04.
| Name | Concentration
|
| pSB4K5 | 60.7 ng/µL
|
| R0011 | 26.8
|
| Name | Sample | 2 buffer | BSA | Enzyme 1 | Enzyme 2 | MilliQ | Total
|
| R0011 [ES] | 50 µL | 6 | 0.6 | EcoRI | 0.2 | SpeI | 0.2 | 3 | 60
|
| pSB4K5 [EP] | 50 | 6 | 0.6 | EcoRI | 0.2 | PstI | 0.2 | 3 | 60
|
| E0240(1) [XP] | 50 | 6 | 0.6 | XbaI | 0.2 | PstI | 0.2 | 3 | 60
|
| E0240(2) [XP] | 50 | 6 | 0.6 | XbaI | 0.2 | PstI | 0.2 | 3 | 60
|
PCR Purification
| Name | Concentration
|
| pSB4K5 [EP] | 39.5 ng/µL
|
| E0240(1) [XP] | 21.8
|
| E0240(2) [XP] | 32.4
|
pSB4K5 [EP] is concentrated 10µL and E0240(1) [XP], E0240(2) [XP] are concentrated 1µL.
Ethanol Precipitation
After ethanol precipitation, we diluted pSB4K5 by 2µL MilliQ
| Name | Vector | Insert 1 | Insert 2 | Ligation High | Total | Incubation
|
| R0011-E0240(1) [pSB4K5] | pSB4K5 [EP] | 1 | R0011 [ES] | 1 | E0240(1) [XP] | 1 | 3 | 15 | 17:30 - 20:20
|
| R0011-E0240(2) [pSB4K5] | pSB4K5 [EP] | 1 | R0011 [ES] | 1 | E0240(2) [XP] | 1 | 3 | 15
|
| Name | Water | MgSO4 | dNTPs | 10xBuffer | Primer VF2 | Primer VR | Template I20260 | KOD plus ver.2 | Total
|
| I20260 (1) | 32µL | 3 | 5 | 5 | 1.5 | 1.5 | 1 | 1 | 50
|
| I20260 (2) | 32 | 3 | 5 | 5 | 1.5 | 1.5 | - | 1 | 50
|
| 94℃ | 2min |
|
| 98℃ | 10s | 30 cycles
|
| 55℃ | 30s
|
| 68℃ | 4min
|
| 4℃ | forever |
|
| Name | Concentration
|
| I20260 | 40.6 ng/µL
|
| Name | Sample volume | 2 buffer | BSA | Enzyme 1 | Enzyme 2 | MilliQ | Total
|
| I20260 [EP] | 45 µL | 6 | 0.6 | EcoRI | 0.2 | PstI | 0.2 | 8 | 60
|
| Name | Concentration | Volume
|
| I20260 [EP] | 74.1 ng/µL | 30
|
I20260 [EP] is concentrated at 7µL
| Vector | Insert | Ligation High | Total | Incubation
|
| I20260 [pSB4K5] | pSB4K5 [EP] | 1 | I20260 [EP] | 1 | 2 | 4 | 20:00-20:30
|
Transformation
| Name | Sample | Competent Cell | Total | Plate | Incubation | Colony
|
| R0011-E0240(1) [pSB4K5] | 1 µL | 20 | 21 | LB (Kan+) | 08/03-08/04 | ○
|
| R0011-E0240(2) [pSB4K5] | 1 | 20 | 21 | ○
|
| I20260 [pSB4K5] | 1 | 20 | 21 | ○
|
Thursday, August 5 By:
Culture and Master Plates
pSB4K5 is inserted RFP generator. We didn't distinguish this inserted parts from low copy plasmid backbone, so self-ligated colony is red. So, white colony is correctly inserted parts.
However, white colonies and green colonies are observed in R0011-E0240(1) [pSB4K5] and R0011-E0240(2) [pSB4K5] plate. We cultured both white and green colonies.
In I20260 [pSB4K5], Many of colonies are red, but green colonies are observed. We cultured green colonies.
| Name | Color | Incubation
|
| R0011-E0240(1) [pSB4K5] (1) | Green Colony | 8/5-8/6
|
| R0011-E0240(1) [pSB4K5] (2) | Green Colony
|
| R0011-E0240(1) [pSB4K5] (3) | White Colony
|
| R0011-E0240(1) [pSB4K5] (4) | White Colony
|
| R0011-E0240(2) [pSB4K5] (1) | Green Colony
|
| R0011-E0240(2) [pSB4K5] (2) | White Colony
|
| R0011-E0240(2) [pSB4K5] (3) | White Colony
|
| R0011-E0240(2) [pSB4K5] (4) | White Colony
|
| I20260 [pSB4K5] (1) | Green Colony
|
| I20260 [pSB4K5] (2) | Green Colony
|
| I20260 [pSB4K5] (3) | Green Colony
|
Sequence
| Name | Concentration
|
| SΔTMD1-E0840(1) A | 28.9 ng/µL
|
| SΔTMD1-E0840(1) B | 25.3
|
| SΔTMD1-E0840(3) A | 26.6
|
| SΔTMD1-E0840(3) B | 24.0
|
As a result, deletion is succeeded, however, point mutation is failed. It is because DpnI is too little to digest all of template DNA.
Friday, August 6
Miniprep
| Name
|
| R0011-E0240(1) [pSB4K5] (1)
|
| R0011-E0240(1) [pSB4K5] (2)
|
| R0011-E0240(1) [pSB4K5] (3)
|
| R0011-E0240(1) [pSB4K5] (4)
|
| R0011-E0240(2) [pSB4K5] (1)
|
| R0011-E0240(2) [pSB4K5] (2)
|
| R0011-E0240(2) [pSB4K5] (3)
|
| R0011-E0240(2) [pSB4K5] (4)
|
| I20260 [pSB4K5] (1)
|
| I20260 [pSB4K5] (2)
|
| I20260 [pSB4K5] (3)
|
Restriction Digestion
| Name | Sample | 2 buffer | BSA | Enzyme 1 | Enzyme 2 | MilliQ | Total
|
| R0011-E0240(1) [pSB4K5] (1) [EP] | 50 µL | 6 | 0.6 | EcoRI | 0.3 | PstI | 0.3 | 2.8 | 60
|
| R0011-E0240(1) [pSB4K5] (2) [EP] | 50 | 6 | 0.6 | EcoRI | 0.3 | PstI | 0.3 | 2.8 | 60
|
| R0011-E0240(1) [pSB4K5] (3) [EP] | 50 | 6 | 0.6 | EcoRI | 0.3 | PstI | 0.3 | 2.8 | 60
|
| R0011-E0240(1) [pSB4K5] (4) [EP] | 50 | 6 | 0.6 | EcoRI | 0.3 | PstI | 0.3 | 2.8 | 60
|
| R0011-E0240(2) [pSB4K5] (1) [EP] | 50 | 6 | 0.6 | EcoRI | 0.3 | PstI | 0.3 | 2.8 | 60
|
| R0011-E0240(2) [pSB4K5] (2) [EP] | 50 | 6 | 0.6 | EcoRI | 0.3 | PstI | 0.3 | 2.8 | 60
|
| R0011-E0240(2) [pSB4K5] (3) [EP] | 50 | 6 | 0.6 | EcoRI | 0.3 | PstI | 0.3 | 2.8 | 60
|
| R0011-E0240(2) [pSB4K5] (4) [EP] | 50 | 6 | 0.6 | EcoRI | 0.3 | PstI | 0.3 | 2.8 | 60
|
| I20260 [pSB4K5] (1) [EP] | 50 | 6 | 0.6 | EcoRI | 0.3 | PstI | 0.3 | 2.8 | 60
|
| I20260 [pSB4K5] (2) [EP] | 50 | 6 | 0.6 | EcoRI | 0.3 | PstI | 0.3 | 2.8 | 60
|
| I20260 [pSB4K5] (3) [EP] | 50 | 6 | 0.6 | EcoRI | 0.3 | PstI | 0.3 | 2.8 | 60
|
==Electrophoresis
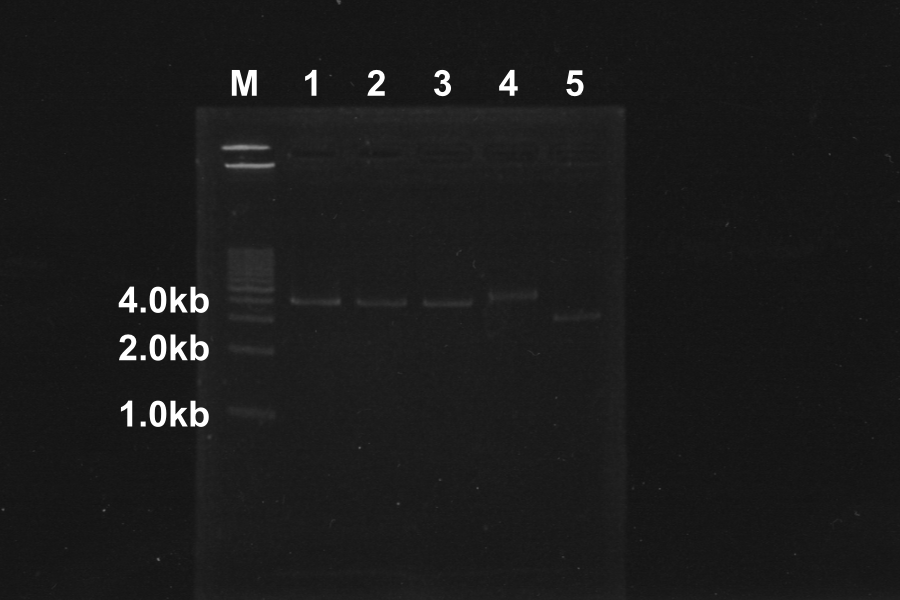
| No. | Name | Length | Results
|
| 1 | I20260 [pSB4K5] (1) [EP] | 960, 4339 |
|
| 2 | I20260 [pSB4K5] (2) [EP] | 960, 4339 |
|
| 3 | I20260 [pSB4K5] (3) [EP] | 960, 4339 |
|
| 4 | R0011-E0240(1) [pSB4K5] (1) [EP] | 980 3378 | ○
|
| 5 | R0011-E0240(1) [pSB4K5] (2) [EP] | 980 3378 | ○
|
| 6 | R0011-E0240(1) [pSB4K5] (3) [EP] | 980 3378 | }
|
| 7 | R0011-E0240(1) [pSB4K5] (4) [EP] | 980 3378 | }
|
| 8 | R0011-E0240(2) [pSB4K5] (1) [EP] | 980 3378 | ○
|
| 9 | R0011-E0240(2) [pSB4K5] (2) [EP] | 980 3378 | }
|
| 10 | R0011-E0240(2) [pSB4K5] (3) [EP] | 980 3378 | }
|
| 11 | R0011-E0240(2) [pSB4K5] (4) [EP] | 980 3378 | }
|
| 12 | I20260 [pSB4K5] (1) [EP] | 960, 4339 | ○
|
| 13 | I20260 [pSB4K5] (2) [EP] | 960, 4339 | ○
|
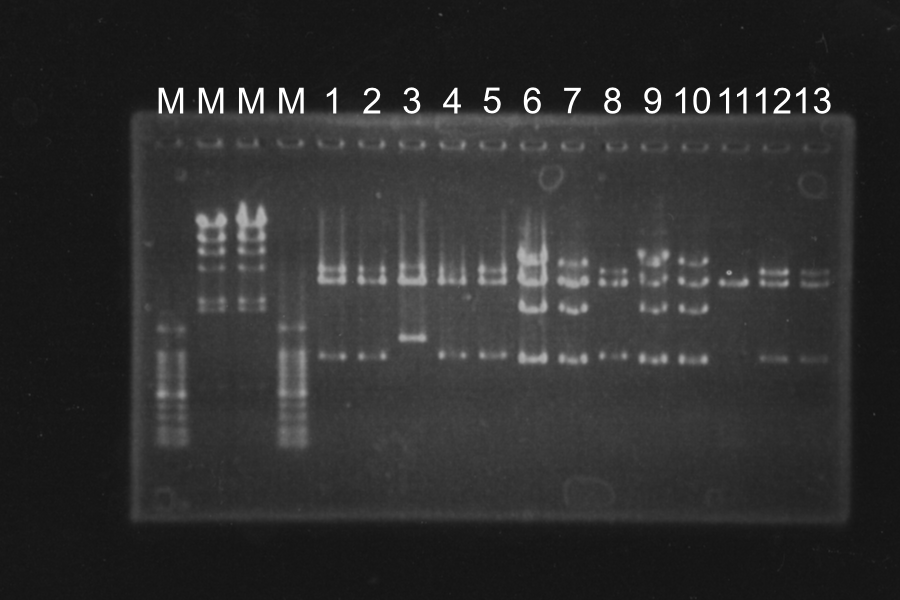 White colonies are not inserted R0011 but its vector. Top10 we used are deleted Lac operon. Then, correctly inserted parts is green because of the lack of lacI gene.
White colonies are not inserted R0011 but its vector. Top10 we used are deleted Lac operon. Then, correctly inserted parts is green because of the lack of lacI gene.
Error PCR (Retry)
| Name | Water | MgSO4 | dNTPs | 10xBuffer | Primer VF2 | Primer VR | Template SSam7,ΔTMD1-E0840 failed (50ng/µL) | KOD plus ver.2 | Total
|
| SSam7,ΔTMD1-E0840 (1) | 32 | 3 | 5 | 5 | 1.5 | 1.5 | 1 | 1 | 50
|
| SSam7,ΔTMD1-E0840 (2) | 32 | 3 | 5 | 5 | 1.5 | 1.5 | 1 | 1 | 50
|
| 94℃ | 2min
|
| 98℃ | 10s | 25 cycles
|
| 68℃ | 4min
|
| Add DpnI 2µL
|
| Incubate | 1h
|
| 4℃ | forever |
|
Transformation
| Name | Well | Sample | Competent Cell | Total | Plate | Incubation | Colony
|
| SSam7,ΔTMD1-E0840 (1) | - | 4 µL | 50 | 54 | LB (Kan+) | 08/06-08/09 | ○
|
| SSam7,ΔTMD1-E0840 (2) | - | 4 | 50 | 54 | ○
|
| I20260 | 2-17-F | 2 | 50 | 52 | ○
|
| 2-I-5 | 2 | 50 | 52 | LB (Amp+) | ○
|
Monday, August 9 By: Wataru, Tomonori, Ken, Takuya
Miniprep
| Name | concentration
|
| I20260 [pSB4K5] | 116.2 ng/µL
|
| R0011-E0240 [pSB4K5] | 146.6
|
Transfotrmation
| Sample | Sample | Competent Cell | Total | Plate | Incuvation | Results
|
| I20260 [pSB4K5] | 2 µL | KRX | 50 | 52 | LB (Kan+) | 08/09 18:00-08/10 12:00 | ○
|
| R0011-E0240 [pSB4K5] | 2 | KRX | 50 | 52 | ○
|
Restriction Eigestion and Ethanol Precipitation
To use R0011 for next ligation, we digested it by EcoRI and PstI
| Name | Sample | 10x Buffer | BSA | Enzyme 1 | Enzyme 2 | MilliQ | Total | Incubation
|
| R0011 [EP] | 50 | 6 | 0.6 | EcoRI | 0.5 | PstI | 0.5 | 2.4 | 60 | At 37℃ 08/09 16:20-18:20
|
After restriction enzyme digestion, we did ethanol precipitation.
Ligation and Transformation
| Sample | Competent cell | Total | Plate | Incuvation | Colony
|
| R0011 [pSB4K5, KRX] | 2 µL | KRX | 50 | 52 | LB (Kan+) | 08/09 20:00-08/10 09:00 | ○
|
| R0011 [pSB4K5</partrinfo>, C2] | 2 | C2 | 50 | 52 | ○
|
====Tuesday, August 10 By: Wataru, Tomonori, Ken, Fumitaka
Culture
Cultured I20260 [pSB4K5, R0011-E0240 [pSB4K5], R0011 [pSB4K5, KRX], and R0011 [pSB4K5</partrinfo>, C2].
Minprep
| Name | Concentration
|
| SSam7,ΔTMD1-E0840 (1-1) | 9.9 ng/µL
|
| SSam7,ΔTMD1-E0840 (1-2) | 27.3
|
| SSam7,ΔTMD1-E0840 (2-1) | 43.2
|
| SSam7,ΔTMD1-E0840 (2-2) | 34.7
|
Culture and Master Plate
At 37℃ 08/09 18:00-08/10 9:00
Wednesday, August 11 By: Wataru, Naoi, Ken, Takuya
| No. | Medium | Cloud | Incubation
|
| 1 | Kanamycin | ○ | At 37℃, 08/10 20:00-08/11 9:00
|
| Ampicillin | }
|
| 2 | Kanamycin | ○
|
| Ampicillin | ○
|
| 3 | Kanamycin | ○
|
| Ampicillin | }
|
| 4 | Kanamycin | ○
|
| Ampicillin | }
|
| 5 | Kanamycin | ○
|
| Ampicillin | }
|
| 6 | Kanamycin | ○
|
| Ampicillin | ○
|
| 7 | Kanamycin | ○
|
| Ampicillin | }
|
Discussion: About sample 1, 3, 4, 5 and 7, lac promoter was correctly inserted in low copy plasmid. About sample 2 and 6, low copy plasmid and vector derived from lac promoter were ligated. We decided to use sample 1 or 3.
Miniprep of R0011 [pSB4K5, C2], SRRz 1', 3'
| Name | Concentration
|
| R0011 [pSB4K5, C2] (1) | 31.2 ng/µL
|
| R0011 [pSB4K5, C2] (3) | 29.9
|
Restriction Digestion and electrophoresis of R0011 [pSB4K5, C2]
| Name | EcoRI | PstI
|
| 1 | 0.2 | -
|
| 2 | - | 0.2
|
| 3 | 0.2 | 0.2
|
| N | - | -
|
| No. | Name | Length | Results
|
| 1 | R0011 [pSB4K5, C2] (1-1) | |
|
| 2 | R0011 [pSB4K5, C2] (1-2) | |
|
| 3 | R0011 [pSB4K5, C2] (1-3) | |
|
| 4 | R0011 [pSB4K5, C2] (1-N) | |
|
| 5 | R0011 [pSB4K5, C2] (2-1) | |
|
| 6 | R0011 [pSB4K5, C2] (2-2) | |
|
| 7 | R0011 [pSB4K5, C2] (2-3) | |
|
| 8 | R0011 [pSB4K5, C2] (2-N) | |
|
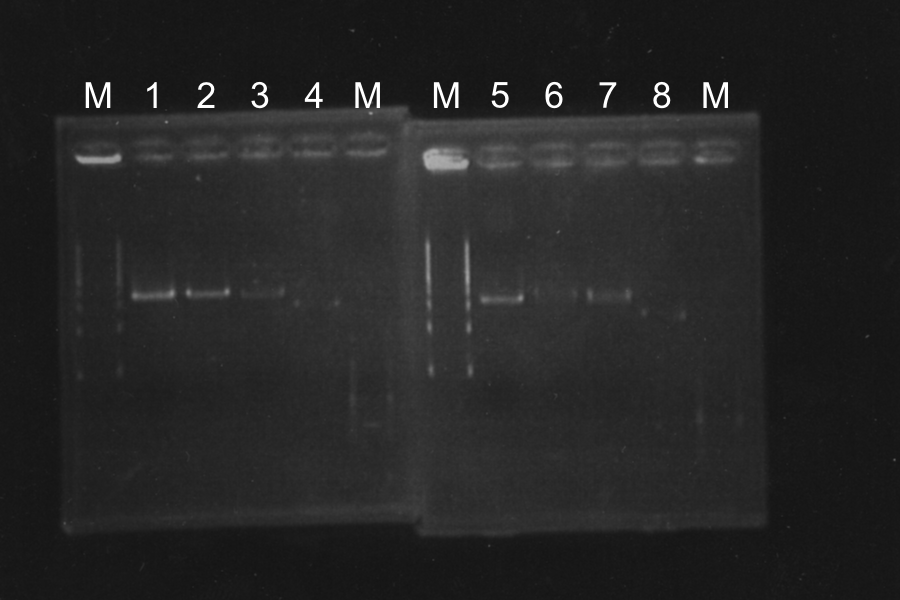 Each enzyme correctly cut samples.
Each enzyme correctly cut samples.
Screening PCR of SRRz
| No. | Name | Results
|
| 1 | None |
|
| 2 | Control B0015 |
|
| 3 | Control J06702 |
|
| 4 | Control B0015 |
|
| 5-24 | SRRz-B0015 | }
|
Marker: Lambda Marker
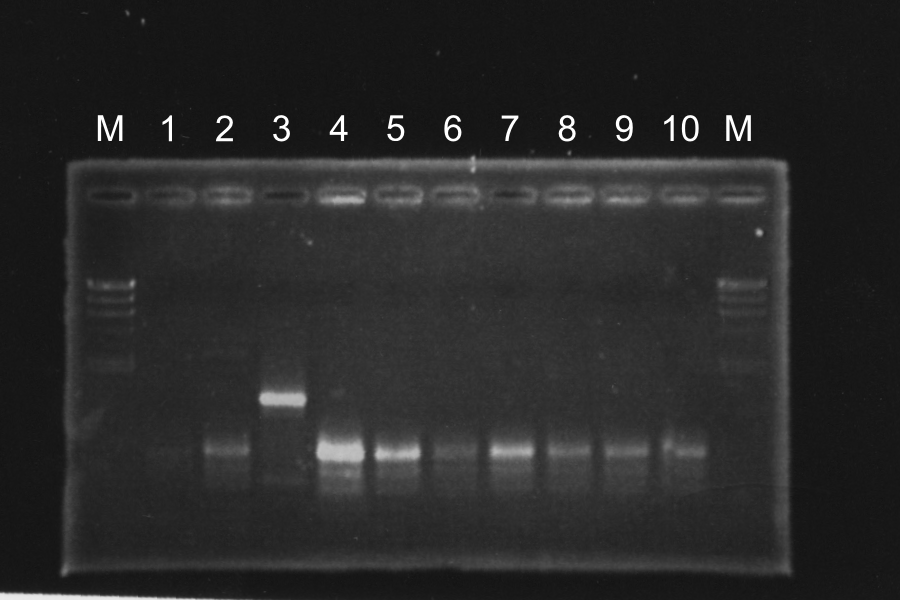
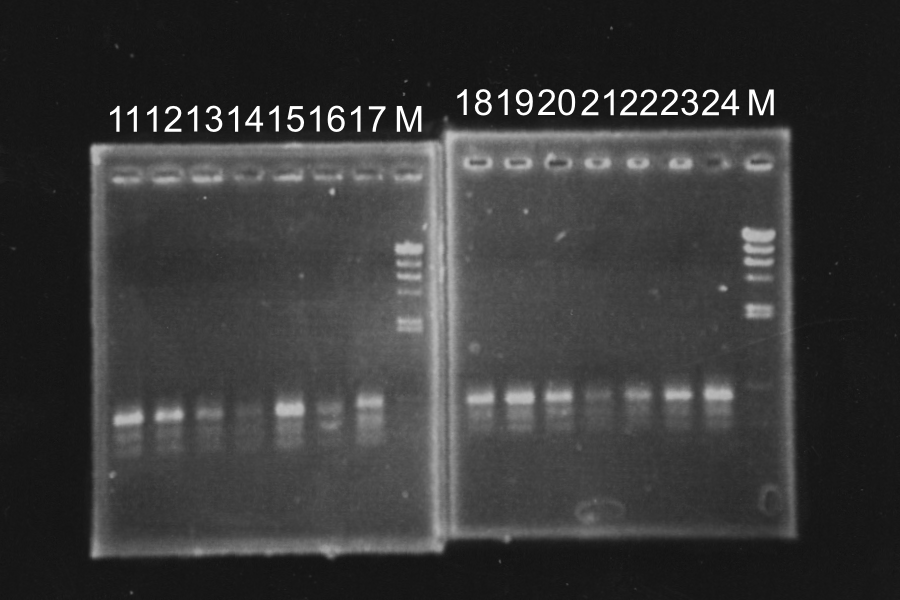 Discussion: All of the sample were self-ligation of DT. SRRz weren't inserted.
Discussion: All of the sample were self-ligation of DT. SRRz weren't inserted.
Thursday, August 12 By: Wataru, Ken
Restriction Digestion and electrophoresis of B0015
| Name | Template | 10xbuffer | 100xbuffer | EcoRI | XbaI 1 | XbaI 2 | SpeI | PstI 1 | PstI 2 | Water | Total
|
| 1 | 3 | 1 | 0.1 | 0.2 | - | - | - | - | - | 5.7 | 10
|
| 2 | 3 | 1 | 0.1 | - | 0.2 | - | - | - | - | 5.7 | 10
|
| 3 | 3 | 1 | 0.1 | - | - | 0.2 | - | - | - | 5.7 | 10
|
| 4 | 3 | 1 | 0.1 | - | - | - | 0.2 | - | - | 5.7 | 10
|
| 5 | 3 | 1 | 0.1 | - | - | - | - | 0.2 | - | 5.7 | 10
|
| 6 | 3 | 1 | 0.1 | - | - | - | - | - | 0.2 | 5.7 | 10
|
| N | 3 | 1 | 0.1 | - | - | - | - | - | - | 5.9 | 10
|
Maker: Lambda, 100bp
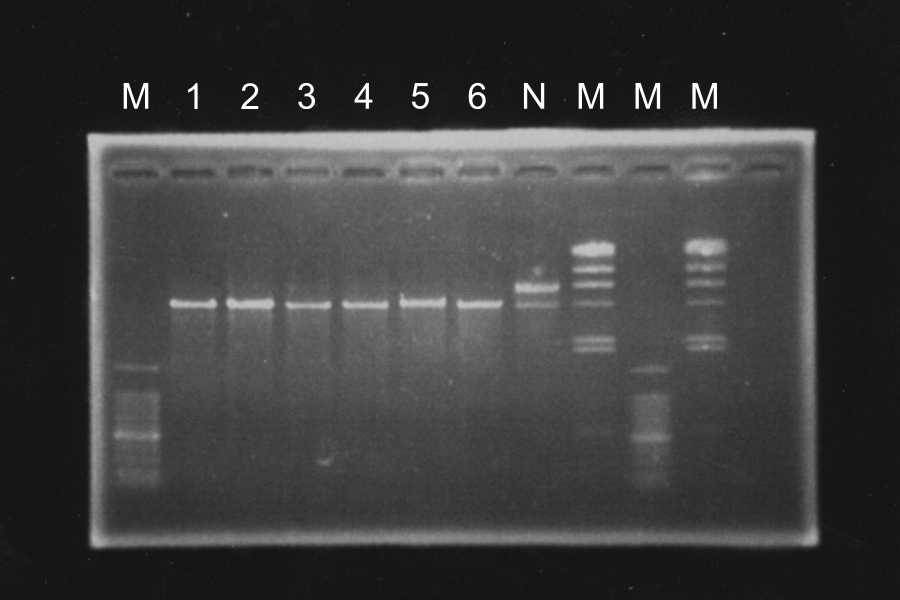 Discussion: Each enzyme correctly cut each sample and was active.
Discussion: Each enzyme correctly cut each sample and was active.
====Thursday, August 19 By: Wataru, Tomo, Ken
Miniprep of SSam7,ΔTMD1-E0840
| Name | Concentration
|
| SSam7,ΔTMD1-E0840 | 29.6 ng/µL
|
Point mutation PCR of SSam7,ΔTMD1-E0840
| Name | Template | 10xbuffer | dNTPs | MgSO4 | Primer Fwd. | Primer Rev. | MilliQ | KOD plus ver.2 | Total
|
| SΔTMD1-E0840 (1) | 1.5 | 5 | 5 | 3 | 1.5 | 1.5 | 31.5 | 1 | 50
|
| SΔTMD1-E0840 (2) | 1.5 | 5 | 5 | 3 | 1.5 | 1.5 | 31.5 | 1 | 50
|
| Control | 1.5 | 5 | 5 | 3 | 1.5 | 1.5 | 32.5 | - | 50
|
| 94℃ | 2min |
|
| 98℃ | 10s | 30cycles
|
| 55℃ | 30s
|
| 68℃ | 3.5min
|
| 4℃ | forever |
|
Restriction Digestion by DpnI from 17:50 to 18:50
Electrophoresis
| Name
|
| SΔTMD1-E0840 (1)
|
| SΔTMD1-E0840 (2)
|
| Control
|
Marker: Lambda, 100bp
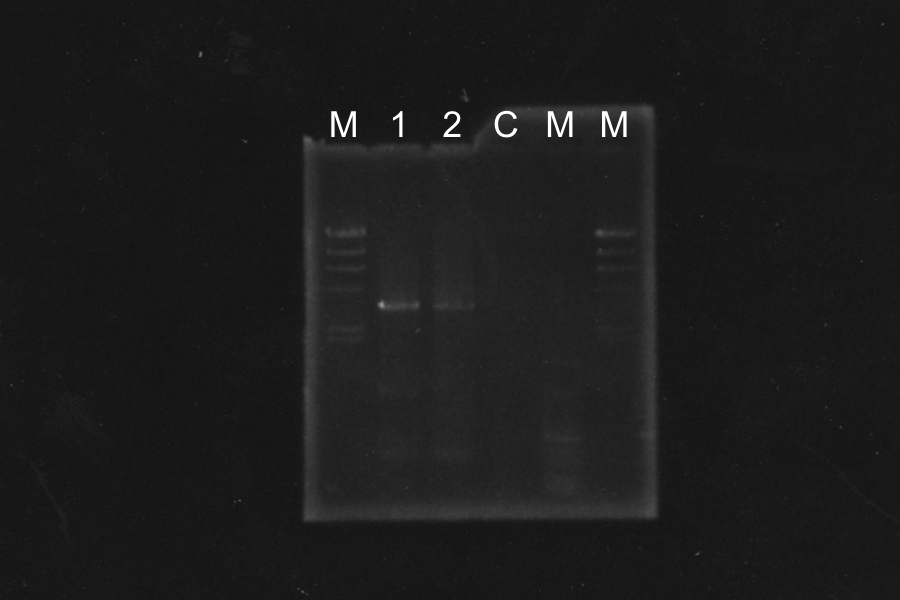
Ligation and Transformation
| Name | Colony
|
| SΔTMD1-E0840 (1) | ○
|
| SΔTMD1-E0840 (2) | ○
|
| Control | }
|
Friday, August 20 By: Wataru, Ken
Making Culture and Master Plate of SΔTMD1-E0840
[[Team:Kyoto/Protocols#Miniprep|Miniprep]
| Name | Concentration
|
| B0015 | 41.1 ng/µL
|
PCR of SRRz
| Name | 10xBuffer | MgS04 | dNTP | Template | Primer Fwd. | Primer Rev. | MilliQ | KOD plus ver.2 | Total
|
| SRRz (1) | 5 µL | 3 | 5 | 5 | F1 | 1.5 | 1.5 | 28 | 1 | 50
|
| SRRz (2) | 5 | 3 | 5 | 5 | F2 | 1.5 | 1.5 | 28 | 1 | 50
|
| SRRz (3) | 5 | 3 | 5 | 5 | F1 | 1.5 | 1.5 | 28 | 1 | 50
|
| SRRz (4) | 5 | 3 | 5 | 5 | F2 | 1.5 | 1.5 | 28 | 1 | 50
|
| SRRz (5) | 5 | 3 | 5 | 5 | F1 | 1.5 | 1.5 | 28 | 1 | 50
|
| SRRz (6) | 5 | 3 | 5 | 5 | F2 | 1.5 | 1.5 | 28 | 1 | 50
|
| 94℃ | 2min |
|
| 98℃ | 10s | 30cycles
|
| 55℃ | 30s
|
| 68℃ | 2min
|
| 4℃ | forever |
|
| Name
|
| SRRz (1)
|
| SRRz (3)
|
| SRRz (5)
|
| SRRz (2)
|
| SRRz (4)
|
| SRRz (6)
|
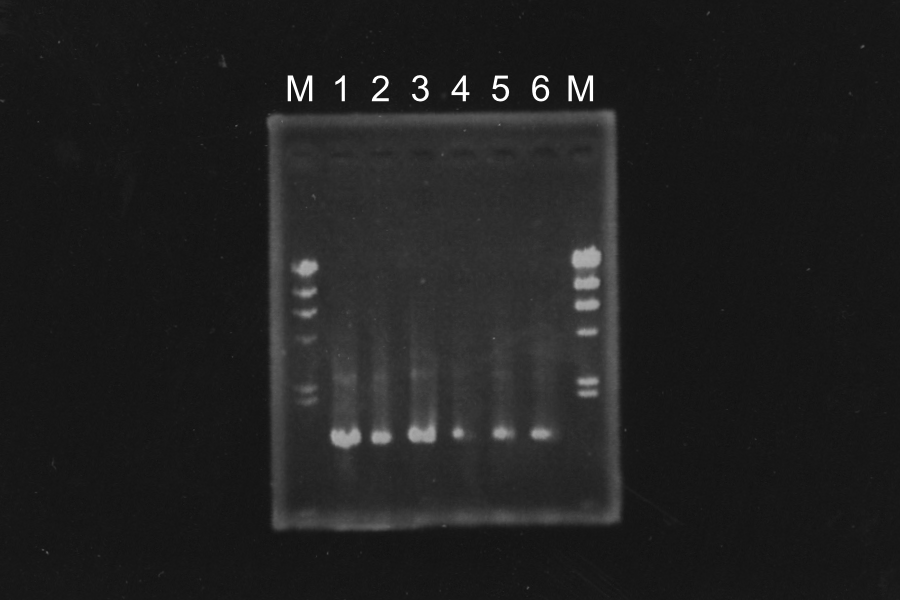 Discussion: Primer F1 might be better than F2, because the bands of 1, 2 and 3 were clearer. We decided to use sample 1 and 3. Their bands were clearer in the three.
Discussion: Primer F1 might be better than F2, because the bands of 1, 2 and 3 were clearer. We decided to use sample 1 and 3. Their bands were clearer in the three.
| Name | Concentration
|
| SRRz (1) | 134.0 ng/µL
|
| SRRz (3) | 69.0
|
| Name | Sample | 10xBuffer | 100xBuffer | EcoRI | XbaI | SpeI | MilliQ | Total | Incubation
|
| B0015 [EX] | 50 µL | 6 | 0.6 | 0.4 | 0.4 | - | 2.6 | 60 | 17:45-18:45
|
| SRRz (1) [EP] | 50 | 6 | 0.6 | 0.4 | - | 0.4 | 2.6 | 60
|
| SRRz (3) [EP] | 50 | 6 | 0.6 | 0.4 | - | 0.4 | 2.6 | 60
|
Purification
| Name | Concentration
|
| SRRz (1) [EP] | 109.0 ng/µL
|
| SRRz (2) [EP] | 110.0
|
| B0015 | 25.5
|
 White colonies are not inserted R0011 but its vector. Top10 we used are deleted Lac operon. Then, correctly inserted parts is green because of the lack of lacI gene.
White colonies are not inserted R0011 but its vector. Top10 we used are deleted Lac operon. Then, correctly inserted parts is green because of the lack of lacI gene.
 Each enzyme correctly cut samples.
Each enzyme correctly cut samples.

 Discussion: All of the sample were self-ligation of DT. SRRz weren't inserted.
Discussion: All of the sample were self-ligation of DT. SRRz weren't inserted.
 Discussion: Each enzyme correctly cut each sample and was active.
Discussion: Each enzyme correctly cut each sample and was active.
 Discussion: Primer F1 might be better than F2, because the bands of 1, 2 and 3 were clearer. We decided to use sample 1 and 3. Their bands were clearer in the three.
Discussion: Primer F1 might be better than F2, because the bands of 1, 2 and 3 were clearer. We decided to use sample 1 and 3. Their bands were clearer in the three.
 "
"