Team:ETHZ Basel/Modeling/Movement
From 2010.igem.org
Modeling of the bacterial movement
Virtual E. lemming vs. Real E. lemming
While the biologists are busy teaching the E. coli how to respond to light, the modelers thought of simulating the light driven E. coli electronically. This will close the loop of our virtual E.lemming and will help us check the imaging pipeline, microscope analysis & joystick control even without actually having the real E. lemming under the microscope.
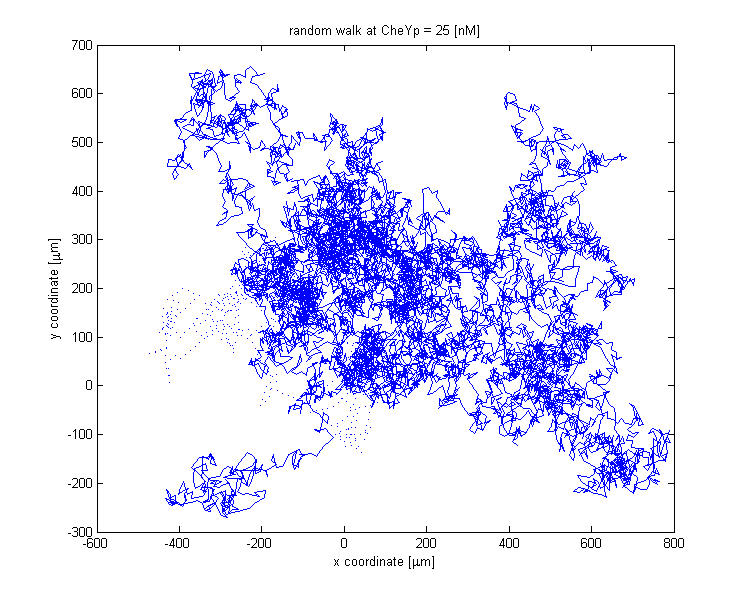
Analogous to the highly complex signal transduction in the Chemotaxis receptor model, there is an equally challenging molecular system on side of the flagella, responsible for the movement of the bacterium. In analyzing this system, we implemented a probabilistic model, meant to realistically simulate the behavior of the cell, by reproducing the main statistical features of the existing empirical data [1]. The chemotactic movement might employ two states, either running or tumbling. The outcome of the flagellar system is under the main influence of CheYp concentration, which is received as an input from the Chemotaxis pathway at every time point of the simulation.
Statistical Objectives
The main target of our algorithm was obtaining very similar statistical results of our simulated E.Coli's movement, when compared to the real E.coli's movement. These statistical results include:
Tumbling Angle
The tumbling angle is the change of direction from run to run. We approximated the empirically observed distribution [1] (left panel) with a Weibull distribution (right panel), with same mean and variance as the empirically observed ones.
Mean Tumbling Length
The mean tumbling length is
Run length
Features of the Movement Model
The Bias
The link between CheYp concentration and the type of movement chosen by the bacterium is called bias and it is formally defined as the fraction of time spent in the running state with respect to the total observation time. Our choice for bias formulation is a nonlinear Hill - type function of the CheYp concentration, with one of the parameters being the CheYp concentration for which a steady - state bias was obtained, well documented from the literature [2]. [NEED FURTHER EXPLANATION!!!]At every time point of the simulation, the cell probabilistically chooses whether to run or to tumble, depending on the bias value. The choice of the running state is equivalent to a change in spatial coordinates, together with a slight change in direction (movement angle), while the choice of the tumbling state corresponds to new movement angle and unchanged spatial coordinates.
Practical Challenges
One of the biggest practical challenges regarding the movement model was obtaining a time - step - invariant behavior of the cell, whose reliability of statistical estimates should be the same, regardless of how often the cell had to choose its future state. In order to achieve this, we opted for a two - state model, in which the probability of running was controlled separately depending on whether the cell was running or tumbling.
Assumptions
The main assumption of our model was that the mean time in tumbling (mean tumbling length) is constant, while the mean time in running (mean running length) is a function of the bias. This assumption was made in accordance with existent published empirical data analyzing the chemotaxis movement. [citation] The movement velocity was also considered constant [citation]
We derived the two central parameters of our model - the running probabilities - by exploiting the mathematical dependency between the mean run/tumbling length and the running/tumbling probabilities:
[derivation of the probability formulas]
!PENDING!
[1] Chemotaxis in Escherichia coli analysed by three - dimensional Tracking. H.C. Berg, D.A.Brown: Nature 239, 500 - 504. 1972
 "
"