Why E.canD?
Two fifths of mankind is suffering from cancer during full life-time, and one fourths dies because of this disease. The rate of people who get cancer increases with age. However, human cannot notice easily about getting cancer or not due to incubation period. Doing so is easier when cancer’s diagnosed at an early stage as treatment is often simpler and more likely to be effective. So finding cancer early is very important.
Half of people are killed by cancer within five years after a diagnosis from doctor, and cancer shortens 15~20 years of lifespan of patients. Another interesting research said that 40% of cancer patients experience severe mental distress; most of them are suffered by pusillanimity. It can make one's life tragedy, not only oneself, but also one's family. Another terrible cause why cancer is severe to us is medical expenses. If someone got a cancer, we will receive huge doctor's fee like hundreds of thousands dollar. It makes terribly huge burden for ordinary family. Unfortunately, someone is sometimes suffering to cancer without any treatment.
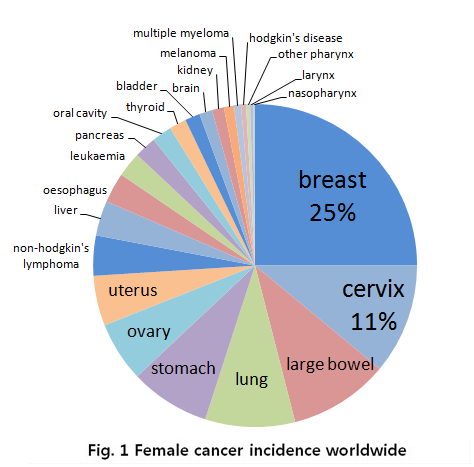
In the case for women, breast cancer is really severe cancer and the most frequent cancer that can find. The following graph shows that a quarter of women who got a cancer have a breast cancer.
There are several methods for cancer diagnosis. One of the widely used methods is imaging test. It is divided into in vivo and in vitro method. In vivo imaging tests are X-ray, MRI, CT, PET, SPCT scan, and Ultrasound, in vitro method is biopsy. A biopsy is a procedure to remove a piece of tissue or a sample of cells from body for laboratory analysis. In addition, the diagnosis methods using bio marker such as RNA and DNA are developed. Bio marker plays a key role in risk assessment, assessing treatment response, and detecting recurrence and the investigation of multiple bio markers may also prove useful in accurate prediction and prognosis of cancers.
We focused on the method with bio maker, especially proteins, which distinguish between normal and cancer cells using expression level difference. Many researchers have chosen antibody to detect these kinds of protein, and it needs special luminescence material to confirm whether antigen (expressed protein) is stacked or not. However it is too complicate and delicate process to attach luminescence substance to antibody and it takes lots of cost and time. In addition, established diagnosis method causes social problem because of gap of demand and supply. For these reasons, we have constructed another accurate and rapid cancer diagnosis system using bacteria. We made bacteria to synthesize fusion antibody and to fix it at the membrane itself and by cognizing antigen bacteria emit fluorescence. We want to establish new diagnosis method with synthetic biology.
Whole Pathway
Flash
- 'E. canD' is spread on the cancer cell.
- E. canD’s antibody combines with the antigen on the cancer cell.
- Antigen-antibody interaction make phosphorylate each receptors.
- JAK autophosphorylation activates transcription factors called STAT1.
- The activated STAT1 dissociate from the receptor.
- The activated STAT1 form dimers.
- The STAT1 dimer regulates GFP gene that attached to GAS elements of promoter.
- 'E.canD' expresses GFP.
Antibodies for Detecting Cancer Cells
Cancer cell has distinctive protein expression pattern. This expression pattern also varies on the cell line of cancer cell. For example CD15 and CD30 proteins are OVEREXPRESSED on Hodgkin’s disease, kind of lymphoma, and CEA is over-expressed on adenocarcinoma cell. These protein expression patterns also vary on the cell line of cancer. For example breast cancer cell has different protein expression pattern; over-expression of Estrogen receptor, Progesterone receptor and KI67 of MCF-7 cell line and over-expression of Human Epidermal Growth Factor receptor, KI67 of AU-56 or SK-BR-3 cell line. Therefore, it allows us to know not only the existence of cancer, but also the type of cell line to identify the protein expression pattern of biopsy sample. We selected Estrogen Receptor and HER2 for our project to make a diagnosis the breast cancer. The reason why we selected these proteins is that they are differently over-expressed. Comparing over-expression pattern of these proteins can diagnose existence of cancer and type of cancer.
Making Biosensor – E. coli Cancer Detector
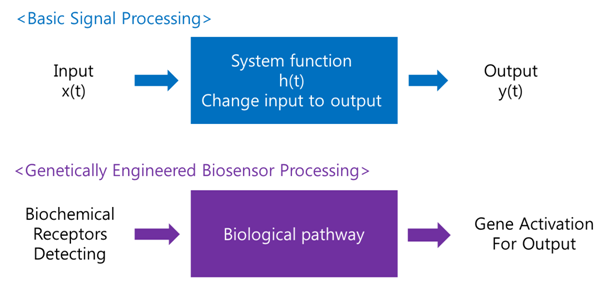
A biosensor is a device for the detection of an analyst that combines a biological component with a physicochemical detector component.[1] In order to make biosensor with genetically engineered organism, we should consider the following basic sensing process.
Receive input -> Process -> Extract output
The ‘Receive input’ part can be alternated by represented receptor, the ‘Process’ part can be signaling pathway which changes input to output, and the ‘Extract output’ part can be something easily detectable by human. Nowadays, pigments or fluorescence proteins are usually used for output, and in order to express these substances, gene expression is made constant use in genetically modified organism. Thus, all engineered biosensors with organisms should be modeled on (consist of) 1) biochemical receptors which can activate gene expression, 2) biological pathway, and 3) gene activation for output.
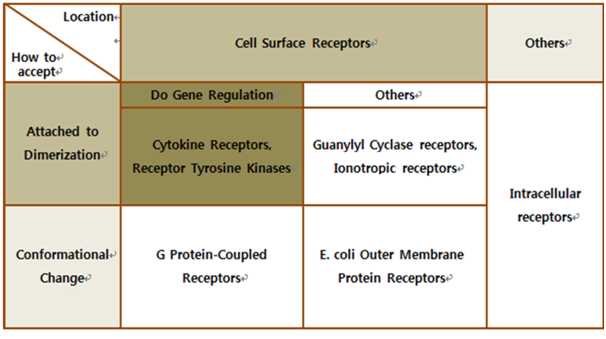
Biochemical Receptors
The upper table represents classification of all biological receptors.
First of all, since our genetically engineered machine has to detect on cancer cell directly, we should select cell surface receptors. Next, we should consider using fusion-antibody-receptors. We used gene combination simply (add antibody to biological receptor without original receipt part). If the receptor has conformational change property when it accepts input signal, we guess that our fusion antibody receptor cannot operate well because antibodies characteristics (antigen size, antibody size, etc.) are different. They don’t have special receipt part in a whole receptor, of course, so whole receptor is important receipt part; we cannot create fusion-antibody-receptor with this protein anyway. Therefore, we choose ‘Dimerization’ receptor. Most of these receptors have Immunoglobulin-like receptor part which can be alternated by antibodies easily. Also, since biosensors should activate gene expression, we selected gene-regulation related receptors.
Finally we choose cytokine receptors and receptor tyrosine kinases. Among them, interleukin-6 alpha receptor was selected.
Fusion Antibody Receptors
The Fusion Antibody-Receptor is a protein which receptor genetically combines with a specific antibody. The following figures show what the fusion antibody-receptor is for details.
The fusion antibody-receptor is designed by simple gene modification. We altered the cancer detectable antibody single chain instead of the cytokine receptor’s antigen acceptable part (Immunoglobulin-like parts). Light chain part of the antibody – Linker – Heavy chain part of antibody is enough. (There are some evidences to success making fusion antibody-receptor with this method in some experiments. [4]) Then in order to express the receptor on outer membrane of E. coli, we change the original signal peptide to Omp(Outer membrane protein) signal peptide of E. coli. Surely the receptor should satisfy our standard for biosensor.
Signal Pathway : JAK-STAT Pathway
There are many signal pathways from plasma membrane receptor to the gene regulation. Figure B is an example of these pathways, MAP kinase pathway linked with RAS pathway. In this pathway,
- With existence of signal molecule, receptor kinases dimerize themselves and phosphorylate each other.
- This phosphorylated receptor dimer is cognized by Grb2 and recruit SOS protein, the Guanine nucleotide Exchange Factor(GEP)
- Recruited SOS protein substitute as GDP of Ras protein with GTP.
- Ras protein with GTP changes its conformation and activate its own kinase activity(Ras pathway so far)
- Ras kinase activate the MAP kinase kinase kinase, MAP KKK(also called as Raf protein) with phosphorylation
- MAP KKK activates the MAP kinase kinase, MAP KK(also called as Mek) with phosphorylation
- MAP KK activates the MAP kinase, MAP K(also called as Erk) with phosphorylation
- Activated MAP Kinase phosphorylates many cytosol proteins.
- For example, if Jun TF is phosphorylated by MAP K, phosphorylated Jun go inside of nucleus and activate the transcription to bind to DNA. (MAP pathway so far)
But to port this signal pathway into the E. coli is not easy because this pathway is related with too many proteins, while JAK-STAT pathway is easy to port because it doesn’t have many participating proteins. The signal transduction step of JAK-STAT pathway as follows.
- Cytokine binds to the cytokine receptor with JAK kinase.
- Kinases form dimer and are phosphorylated by JAK.
- These phosphorylated tyrosine residue is cognized by STAT protein.
- STAT which cognize phosphorylated tyrosine phosphorylate other STAT protein.
- Two phosphorylated STAT protein form dimer and go inside to the nucleus and activate near gene to bind to the GAS motif.
Because JAK-STAT pathway have only 5 steps while RAS-MAP pathway require 9 steps, we decide to port JAK-STAT pathway to the E. coli.
Gene Activation for Output
Inside the nucleus the active STAT dimer binds to cytokine inducible promoter regions of genes containing gamma activated site (GAS) motif and activate transcription of cytokine inducible genes.[5] The STAT protein can be dephosphorylated by nuclear phosphatases which lead to inactivation of STAT and the transcription factor becomes transported out of the nucleus by exporting crm1/RanGTP. Using this pathway, we altered the green fluorescent protein (GFP) gene instead of cytokine inducible genes. So, we could show GFP expression through the previous whole pathway.[Fig. 5]
References
- Fig 1. Female cancer incidence worldwide, MRC, National Institute for Medical Research
- http://www.nimr.mrc.ac.uk/virology/doorbar/fig1/
- Fig 2. Nature Biotechnology 22, 6 - 7 (2004)
- http://www.nature.com/nbt/journal/v22/n1/full/nbt0104-6.html
- Fig 3.Bon cancer symptoms (2008)
- http://bonecancersymptoms.org/
- Fig 4. Charlotte Mulvihill, author. Licensed for use, ASM MicrobeLibrary
- http://vivo.cornell.edu/individual/vivo/individual847
- Fig 5. RCSB Protein Database “Crystal structure of the hexameric human IL-6/IL-6 alpha receptor/gp130 complex ”
- http://www.rcsb.org/
- Fig 6. Roberto, Proteína fluorescente verde – história e perspectivas, Química de Produtos Naturais (2009)
[1] International Union of Pure and Applied Chemistry. "biosensor". Compendium of Chemical Terminology Internet edition.
[2] Uniprot Database “Interleukin-6 receptor subunit alpha”; http://www.uniprot.org/uniprot/P08887
[3] Lichtman, Andrew H.; Abbas, Abul K. Cellular and molecular immunology (5th ed.). (2003). Philadelphia: Saunders. ISBN 0-7216-0008-5.
[4] SR Roffler1,6, H-E Wang2,6, H-M Yu2, W-D Chang3,4, C-M Cheng3,4, Y-L Lu5, B-M Chen1
and T-L Cheng3,4. A membrane antibody receptor for noninvasive imaging of gene expression. Gene Therapy. Nature (2005)
[5] Martine I. Darville, Ye-Shih Ho and Decio L. Eizirik, NF-B Is Required for Cytokine-Induced Manganese Superoxide Dismutase Expression in Insulin-Producing Cells, The Endocrine Society(2000)
|
Sub-Menu
abcdefghijklmnopqrstuvwxyz
|
 "
"