Team:ETHZ Basel/Biology/Cloning
From 2010.igem.org
Cloning strategy for the construction of our Biobricks
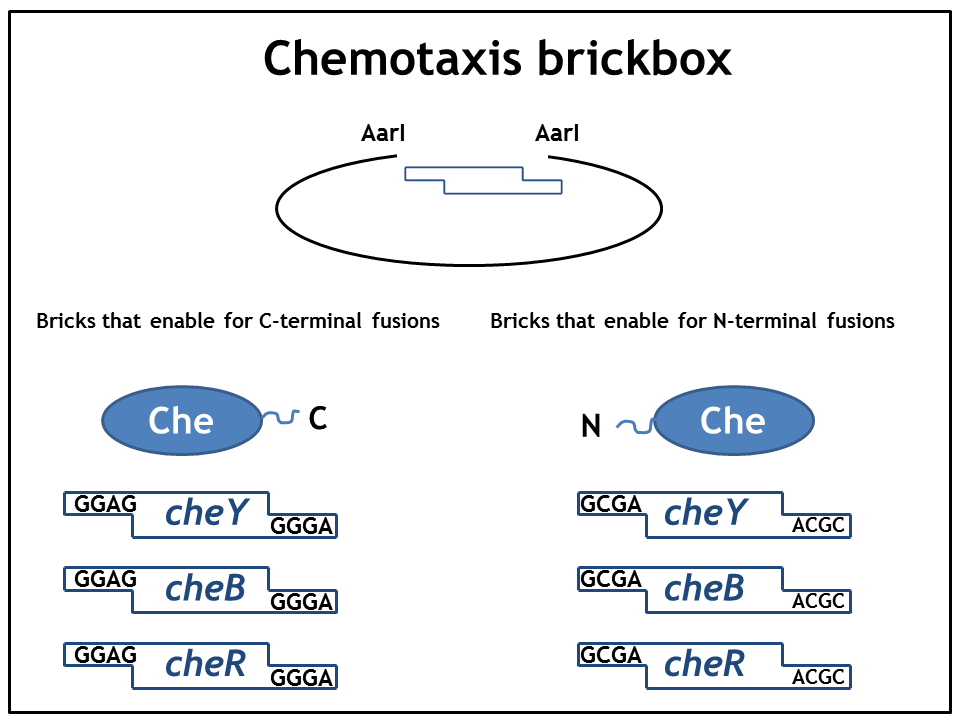
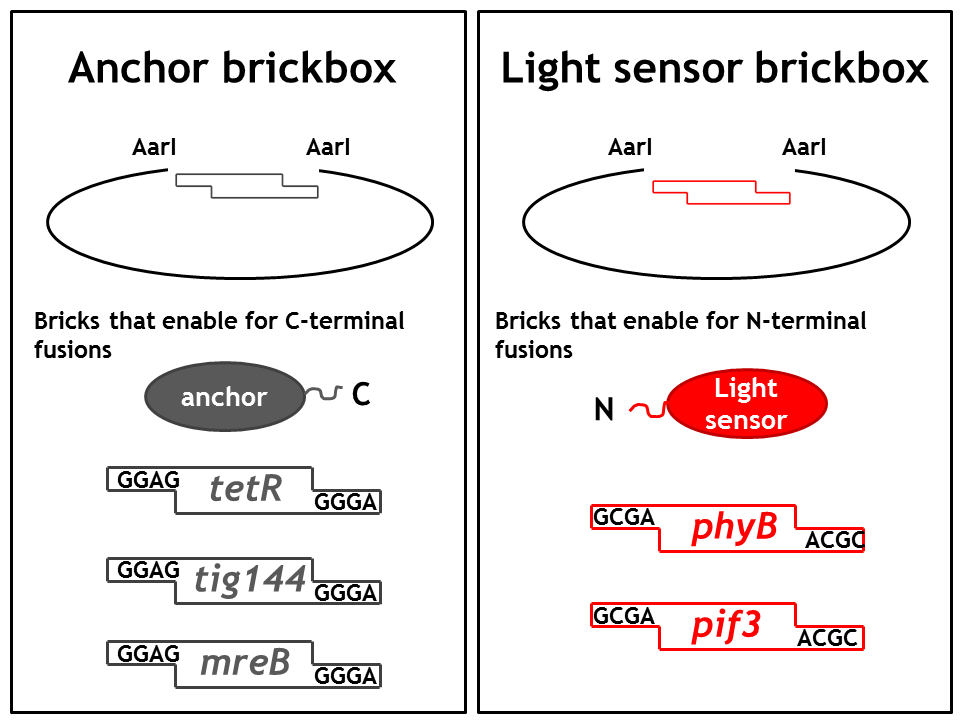
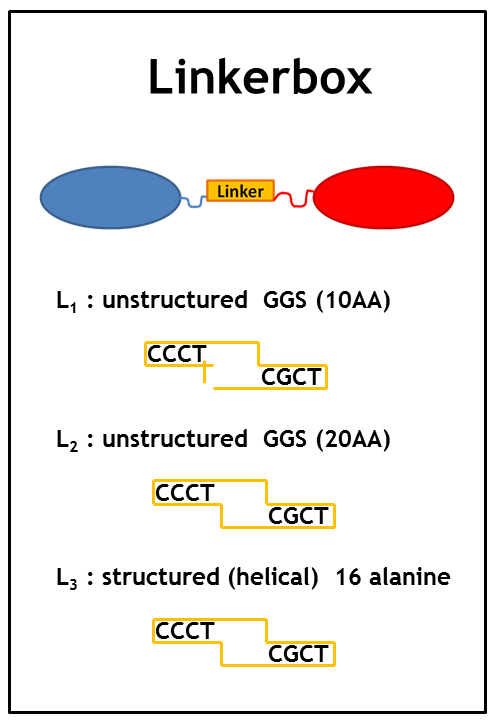
As we planned to generate several fusion proteins with different linkers, we decided to use the cloning strategy BBF RFC28: A method for combinatorial multi-part assembly based on the Type IIs restriction enzyme AarI [1]. The advantage of this strategy is that we can clone up to 3 different inserts into one expression vector simultaneously. In the following section we give an overview of how this was achieved.
1. Step: Construction of brickboxes that enable for the generation of fusion proteins
Parts were generated by PCR using primers specified in the BBF RFC28 manual and subcloned into the storage vector pSEVA132 (Victor de Lorenzo's lab, KanR, pBBR1 ori) by blunt end ligation. pSEVA132 allows for blue white screening, making the generation of the brickbox parts very efficient. Generated parts were verified by AarI digest and sequencing. Due to the presence of rare codons in the sequence of PhyB and Pif3, these two genes were codon optimized and ordered from GeneArt. As the implementation of E. Lemming relies on two fusion proteins (the "anchor-light sensor" and "light sensor-che protein" fusion), two expression vectors - so called working vectors- were constructed, which enable for the simultaneous expression of the two fusion proteins. Working vector 1 is a derivative of pSEVA132 conferring resistance to ampicillin and replicating with ori pBB1. Working vector 2 is a derivative of pSEVA421 expressing a spectinomycin resistance cassette and replicating with ori RK2. The gene for the repressor AraC and the corresponding ParaBAD promotor/operator sites were introduced into both vectors followed by an insert flanked by AarI-recognition sites. Digest with AarI releases the insert and generates a vector with assembly compatible overhangs.
2. Step: Assembly of fusion proteins using the generated brickboxes
The following image illustrates the assembly of a fusion protein. In the section "Implementation" the fusion proteins are described, which were actually created to implement E. lemming. The decision which fusion system to implement was taken due to the in silico predicted requirements for a working system.
Linking BBF RFC28 to the Tom Knight's [http://dspace.mit.edu/handle/1721.1/21168 original assembly standard]:
General scheme for the design and construction of Tom Knight's BBF-compatible fusion proteins
BBF RFC28 - A method for combinatorial multi-part assembly based on the Type IIs restriction enzyme AarI - is a very efficient method to construct fusion proteins. Unfortunately it is not compatible with the general iGEM requested Tom Knight's original assembly standard as the fusion protein will not readily contain the required prefix and suffix sequences. During our project we learned a lot about the cloning of fusion proteins using BBF RFC28 and especially how to make it work efficiently. We would like to share our experience here, especially as we worked out a general design method for the easy construction of Tom Knight's original assembly standard and BBF RFC28 compatible working vectors (the expression vector that is finally used for the assembly) that can be used for the final assembly step. Our vector design allows in addition for a GFP (or any other reporter gene)-mediated screening for fusion protein containing clones. In general the working vector needs to contain an insert flanked by AarI-sites. These cleavage sites need to be designed such that digestion with AarI releases the insert and leaves a vector backbone with 5` overhangs compatible with the BBF RFC 28 standard. To isolate 100% cleaved vector backbone for efficient assembly, Insert, cleaved vector and uncut vector can be separated on a 1% agarose gel. This is crucial as AarI does not cleave every cleavage site with high efficiency and alarge fraction will be uncut vector. By choosing as insert a constitutively expressed GFP (it can be any other constitutively expressed reporter gene) positive clones containing the assembled protein can be distinguished from clones containing the original insert (traces of uncut vector might still be present in the cleaved vector fraction) by green/white screening.
References
[1] [http://dspace.mit.edu/handle/1721.1/46721 BBF RFC 28: A method for combinatorial multi-part assembly based on the Type IIs restriction enzyme AarI]
 "
"