Team:Stockholm/4 October 2010
From 2010.igem.org
(Difference between revisions)
(→Andreas) |
(→Andreas) |
||
| Line 21: | Line 21: | ||
====ON cultures==== | ====ON cultures==== | ||
*3 ml LB, 30 °C | *3 ml LB, 30 °C | ||
| - | *# A | + | *# A (BL21 pEX.nLMWP⋅SOD⋅His) |
| - | *# A | + | *# A (BL21 pEX.nTra10⋅SOD⋅His) |
===Transfer of nCPP⋅SOD⋅His.RBS.yCCS operon to pEX=== | ===Transfer of nCPP⋅SOD⋅His.RBS.yCCS operon to pEX=== | ||
| Line 47: | Line 47: | ||
====ON cultures==== | ====ON cultures==== | ||
*5 ml LB, 37 °C, 250 rpm | *5 ml LB, 37 °C, 250 rpm | ||
| - | ** pEX.nTAT⋅SOD⋅His.RBS.yCCS 2: A | + | ** pEX.nTAT⋅SOD⋅His.RBS.yCCS 2: A |
| - | ** pEX.nTAT⋅SOD⋅His.RBS.yCCS 3: | + | ** pEX.nTAT⋅SOD⋅His.RBS.yCCS 3: B |
| - | ** pEX.nTra10⋅SOD⋅His.RBS.yCCS 1: | + | ** pEX.nTra10⋅SOD⋅His.RBS.yCCS 1: B |
| - | ** pEX.nTra10⋅SOD⋅His.RBS.yCCS 2: | + | ** pEX.nTra10⋅SOD⋅His.RBS.yCCS 2: B |
| - | ** pEX.nLMWP⋅SOD⋅His.RBS.yCCS 2: | + | ** pEX.nLMWP⋅SOD⋅His.RBS.yCCS 2: B |
| - | ** pEX.nLMWP⋅SOD⋅His.RBS.yCCS 3: | + | ** pEX.nLMWP⋅SOD⋅His.RBS.yCCS 3: B |
===Verification of pSB1x3 plasmids=== | ===Verification of pSB1x3 plasmids=== | ||
Revision as of 14:29, 5 October 2010
Contents |
Andreas
Transfer of pEX.nCPP⋅SOD⋅His to BL21
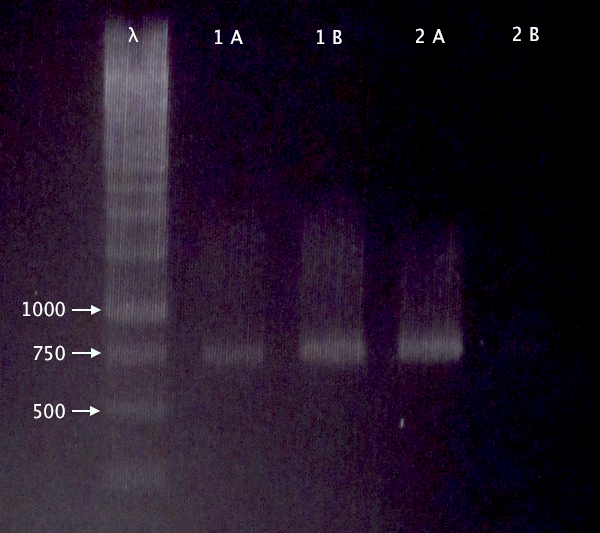
Gel verification
Re-run of 2/10 BL21 samples
- BL21 pEX.nLMWP⋅SOD⋅His: A & B
- BL21 pEX.nTra10⋅SOD⋅His: A & B
1 % agarose, 120 V
Expected bands:
- 744 bp
- 765 bp
Results
- Both clones verified
- Clone A verified; weak band for clone B
ON cultures
- 3 ml LB, 30 °C
- A (BL21 pEX.nLMWP⋅SOD⋅His)
- A (BL21 pEX.nTra10⋅SOD⋅His)
Transfer of nCPP⋅SOD⋅His.RBS.yCCS operon to pEX
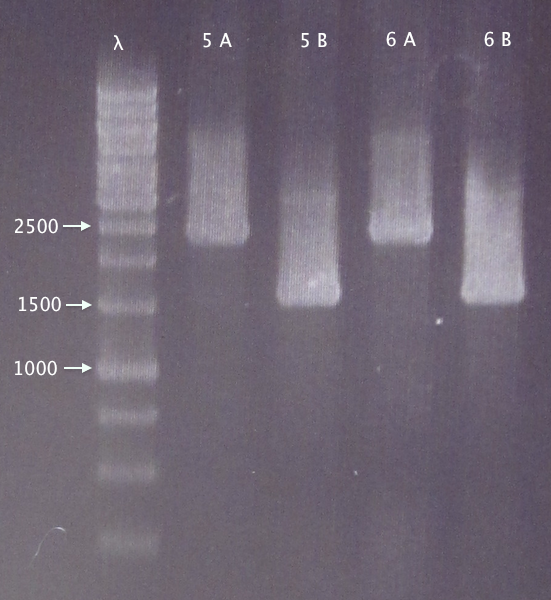
Colony PCR
Picked 2 new colonies of each of the two constructs transformed 30/8:
- 5. pEX.nTra10⋅SOD⋅His.RBS.yCCS 1: A & B
- 6. pEX.nTra10⋅SOD⋅His.RBS.yCCS 2: A & B
Standard colony PCR settings
- Elongation time: 2:00
Gel verification
0.8 % agarose, 100 V
Expected bands:
- 5. 1553 bp
- 6. 1553 bp
Results
- 5. Relevant band for clone B; too large insert (double?) for clone A.
- 6. Relevant band for clone B; too large insert (double?) for clone A.
ON cultures
- 5 ml LB, 37 °C, 250 rpm
- pEX.nTAT⋅SOD⋅His.RBS.yCCS 2: A
- pEX.nTAT⋅SOD⋅His.RBS.yCCS 3: B
- pEX.nTra10⋅SOD⋅His.RBS.yCCS 1: B
- pEX.nTra10⋅SOD⋅His.RBS.yCCS 2: B
- pEX.nLMWP⋅SOD⋅His.RBS.yCCS 2: B
- pEX.nLMWP⋅SOD⋅His.RBS.yCCS 3: B
Verification of pSB1x3 plasmids
Due to some strange growth results with our stock plasmids (pSB1x3.BBa_J04450), I decided to verify their antibiotic resistance. Restreaked clones of the following plasmids (w/ BBa_J04450 inserts) onto Amp 100, Km 50 and Cm 25 plates:
- pSB1A3
- pSB1C3
- pSB1K3
- pSB1AC3
- pSB1AK3
Assembly of His⋅SOD⋅cCPP constructs
Continued from 1/10
Received cCPPs (cTra10, cTAT and cLMWP) in pSB1C3 plasmids, digested with EcoRI and NgoMIV, from Johan.
Ligations
- Vectors:
- Dig pSB1C3.cTra10 E+N
- Dig pSB1C3.cTAT E+N
- Dig pSB1C3.cLMWP E+N
- Insert: Dig pMA.His⋅SOD E+A
| 1 | 2 | 3 | |
|---|---|---|---|
| 10X T4 Ligase buffer | 2 | 2 | 2 |
| Vector DNA | 1 | 1 | 1 |
| Insert DNA | 5 | 5 | 5 |
| dH2O | 11 | 11 | 11 |
| T4 DNA ligase | 1 | 1 | 1 |
| 20 μl | 20 μl | 20 μl |
- Incubation: 22 °C, 15 min
Transformations
- pSB1C3.His⋅SOD⋅cTra10
- pSB1C3.His⋅SOD⋅cTAT
- pSB1C3.His⋅SOD⋅cLMWP
- Standard transformation
- 1 μl
- Cm 25
 "
"