Team:DTU-Denmark/Project
From 2010.igem.org
| Line 153: | Line 153: | ||
<td width="650 px"> | <td width="650 px"> | ||
<h1>Background</h1> | <h1>Background</h1> | ||
| - | <h3>What is Synthetic Biology</h3> | + | <h3>What is Synthetic Biology?</h3> |
<p align="justify">Not many people have heard of Synthetic Biology. Synthetic biology essentially aims to utilize natures tricks to design and build artificial biological systems for engineering purposes as well as a way to get a better understanding of why biological systems are set up as they are. The term “synthetic biology” was first used on genetically engineered bacteria that were created with recombinant DNA technology. Parts from natural biological systems are taken, characterized and simplified and used as a component of a highly unnatural, engineered, biological system. The term was then used when referring to when organic synthesis is used to generate artificial molecules that mimic natural molecules such as enzymes. | <p align="justify">Not many people have heard of Synthetic Biology. Synthetic biology essentially aims to utilize natures tricks to design and build artificial biological systems for engineering purposes as well as a way to get a better understanding of why biological systems are set up as they are. The term “synthetic biology” was first used on genetically engineered bacteria that were created with recombinant DNA technology. Parts from natural biological systems are taken, characterized and simplified and used as a component of a highly unnatural, engineered, biological system. The term was then used when referring to when organic synthesis is used to generate artificial molecules that mimic natural molecules such as enzymes. | ||
There are two types of synthetic biologists: | There are two types of synthetic biologists: | ||
| Line 160: | Line 160: | ||
<li>those who deal with designing and fabricating biological components that do not already exist in the real world. </li> | <li>those who deal with designing and fabricating biological components that do not already exist in the real world. </li> | ||
</ul> | </ul> | ||
| - | Synthetic biology provides us with a new perspective from which we can understand and ultimately utilize life for our own benefits. | + | Synthetic biology provides us with a new perspective from which we can understand and ultimately utilize life for our own benefits. [1,2,3] |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 374: | Line 374: | ||
* (Jensen 2004) Ole Nørregaard Jensen, “Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry,” Current Opinion in Chemical Biology 8, no. 1 (February 2004): 33-41. | * (Jensen 2004) Ole Nørregaard Jensen, “Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry,” Current Opinion in Chemical Biology 8, no. 1 (February 2004): 33-41. | ||
| + | * http://syntheticbiology.org/FAQ.html | ||
| + | * http://www.nature.com.globalproxy.cvt.dk/nrg/journal/v6/n7/execsumm/nrg1637.html | ||
| + | * http://www.nature.com.globalproxy.cvt.dk/msb/journal/v2/n1/full/msb4100073.html | ||
Revision as of 13:21, 5 October 2010
| Home | The Team | The Project | Parts submitted | Modelling | Notebook | Blog |
BackgroundWhat is Synthetic Biology?Not many people have heard of Synthetic Biology. Synthetic biology essentially aims to utilize natures tricks to design and build artificial biological systems for engineering purposes as well as a way to get a better understanding of why biological systems are set up as they are. The term “synthetic biology” was first used on genetically engineered bacteria that were created with recombinant DNA technology. Parts from natural biological systems are taken, characterized and simplified and used as a component of a highly unnatural, engineered, biological system. The term was then used when referring to when organic synthesis is used to generate artificial molecules that mimic natural molecules such as enzymes. There are two types of synthetic biologists:
|
Introduction
Overall Goal
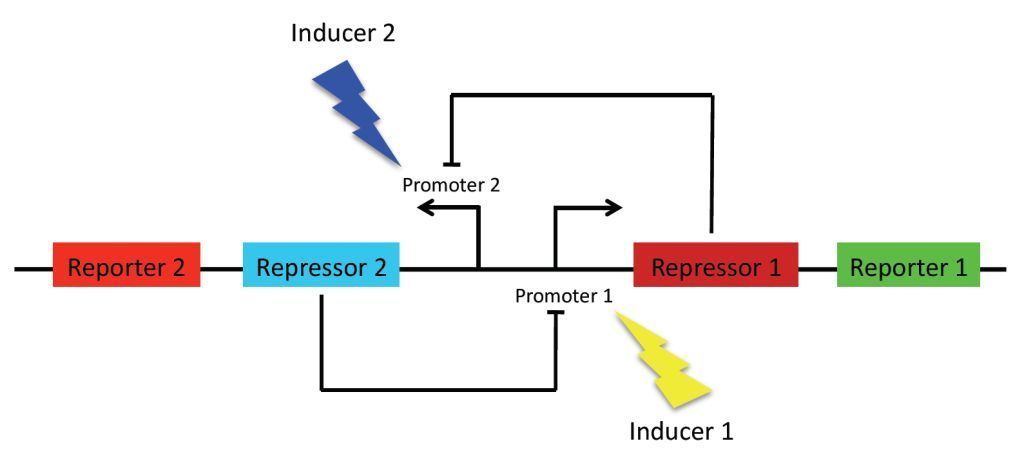
The goal of our project is to enable colonies of E. coli bacteria to transition between production of two different reporter proteins. In our system, switching between states will be induced by exposing the bacteria to light. Each of the states will have a specific frequency associated with it. There are multiple potential applications for biologicals "switches" such as these, this includes the improved control of production of additives in industrial biotechnological processes.
Project Concept
As previously stated, the main goal of our project is to design a bistable switch. The switching between the two states will be controlled by the introduction of two different wavelengths of light, each wavelength responsible for the induction of a different state. As a proof of concept, we’re using fluorescent proteins as reporter genes which makes it easy to observe and characterise the system. In principle, however, any reporter gene can be used.
Our original project concept revolved around using light-receptors to instigate the switch between the two stable state. It was thought that the production of the first reporter protein would be induced by red light (660 nm). At the same time, production of the other reporter will be suppressed by a coexpressed repressor. Conversely, production of the second reporter would be induced by blue light (470 nm). Bistability of the system is achieved by using two repressors which negatively regulate each other’s expression. This enables the system to sustain state without continuous input, i. e. once production of a reporter protein is initiated, it will persist until the system is forced into the other state.
Our project concept has since changed to using two different carbohydrate sources as a means of switching between the two stable states. This means that the state in which the bacteria will be found depends on which one of two carbohydrate sources it was last exposed.
Background
(INTRODUCTION)
Biological switches
what is a biological switch, examples and existing constructions, What can we use it for. what have been build. Many small cuircuits have been constructed. And reviews have been done on also trying to build regulatory function in enzymes. See article (“Designing switchable enzymes” Marc Ostermeier)
Fluorescence Proteins
as we departed in the idea from the terminator screening plasmids described in the partsregistry.org (REFFF) we had a primary focus on fluorescence proteins as our reporter systems. Further we wanted to have high quality data, with a high resolution. We descided on the in-house expertice on using a continous microfermentor system that can measure two fluorescence proteins continuously (biolector) and a flow cytometer, also capable of measuring at two different wave length.
Micro fermentor systems
description of the biolector and a few references
Flow cytometry
Description of flow cytometry a few references
The RNA Polymerase (RNAP)
In synthetic biology when creating the minimal cell XXX factors have been identified as essential for normal function of RNAP, both regarding normal elongation but also in terms of normal termination function. (REFFF) of these XXX have been identified to take part in regulatory function. While the rest is core subunits. Of interest for regulation in terms of termination Anti-termination will be highlighted:
- NusA:
- NusG:
- NusE
- NusB
Due to the construction of the RNAP of many subcomponents and systems, the function of the RNAP can be regulated by only adding or changing one or a few factors. This is the mechanism in the different termination and anti-termination functions described below.
Figure and table containing normal transcription and normal termination and table with sub-part names and explanation.
Termination =
(terminator introduction) Termination can fall into one of two catagories:
- Intrinsic Termination
- Factor-dependent Termination
Intrinsic Termination
Intrinsic Termination can be found to occur at defined template sequences, usually a region of hyphenated inverted sequence symmetry followed by a run of T residues. Termination through intrinsic terminators is stimulated by additional factors, e.g. NusA. Termination occurs due to the stem-loop structure formed by the base-pairing of mRNA with itself caused by inverted sequence symmetry, followed by the run of T residues. The NusA protein causes the RNA-p complex to temporarily stall at the stem-loop structure, when this is followed by a poly-A tail, the RNA-DNA duplex is destabilized. This causes the RNA-p to dissociate from the DNA, thereby terminating transcription. Termination function step-by-step. Factor dependent Termination
Factor dependent Termination
Factor-dependent Termination occurs due to events that are not directly related to transcription, such as the release of ribosomes from nascent transcript or DNA damage. One such host termination factor is Rho, which acts on many sites along the bacterial chromosome. (( ??MFD, is a host termination factor that is responsible for releasing RNA-p stalled at sites of UV-induced DNA lesions. ??)) rho-dependent termination is characterized by not having a specific hairpin structure involved in the termination. The termination thus happens whendue to XXXXXX, and what have been found of the termination site any commen sequences or consensus ????????????????? the function of rho dependent termination, have been shown to be affected by XXXX. The rho binding sites on the mRNA, have been identified from XXbp to XXXbp of stream of the termination site. Rho-termination is thus an example of the more complex termination regulation that is not fully understood and can be very difficult to define and use for engineering purposes. Thus for a more defined anti-termination system the lambda N-protein system and the interaction with the nut-site in the phage genome ????? is a more defined system. Recent research have found out that the Rho termination
Phages
What do they need to be succesfull? How stable have the function been proved to be. Examples of known phage regulation systems. Lambda, p21, P22, gifsy1,2,3,. What does the genomes contain of proteins, mRNA, binding sequences, ANJA??? Identification of mechanisms in regulation, compare with the Chinese paper (hong kong) using cI protein and UV-radiation. Link from phages to anti-terminator and/or Repressor function in phages (next sections)
Phage Repressor System
Maya Lisa anja
Alpha-repressor
The C1-repressor is responsible for repressing transcription of the lytic genes, thereby maintaining the stable lysogenic state. The induction of the lytic state is caused by activated RecA, which stimulates the self-cleavage of the C1-repressor. We will be using the C1-repressor in our system.
Phage Anti-Termination system
(INTRO)
Anti-Termination is the process by which the termination of gene transcription is prevented. Such control of gene transcription can be found in the phage Lambda system. The mechanism is controlled by proteins, such as the lambda N or lambda Q-proteins. The expression of early genes and late genes are both regulated by the anti-termination mechanism, controlled by the lambda N-protein and the lambda Q-protein, respectively. The N-protein is able to suppress transcription termination at both factor-dependent and factor-independent termination sites. N anti-termination is strongly stimulated by the NusA protein. Unlike the N-protein, the Q-protein specifically binds to a DNA sequence immediately upstream of the pR´ promoter.
(FIGURE + FIGURE TEXT)
A more detailed explanation of these anti-termination mechanisms will be posted later on. The mechanism of N-protein nut-site termination have been studied heavily the last years and the current best descriped mechanism have been done in a couple of reviews (XXXXX,XXXX,XXXX) The anti-termination function by introduction of the N-protein (or equivalent) that interacts with nusA and disrupt the termination. Of known systems can be mentioned lambda, p21, p22 FUNCTION XXXXXXXXXXXX shown in the figure below. In the known systems the nut site is placed from XXX bp to XXX bp upstream of the termination steam loop. (REFERENCES !!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!! IMPORTANT - read two gottersman 2010 – crystallography papers)))) figures from papers on antitermination. Few papers descripe and test the the actual needed distance from the termination steam loop. From other systems and mechanism it is known that XXX bp is needed for regulation of RNAP or DNAP. (MOGENS ABOUT REFERENCES AND SYSTEMS SEE MICRO-BIO-TEXT BOOKS_) .
N-protein plasmid
The N protein were isolated from salmonella genomic DNA with specific designed primers. We used the natural occurring RBS site, as a High expression of N have shown non specific anti-termination effect on a global scale on the genome. #References References
nut sites
Severel papers analyse the function of the nut-site. It has been shown that this mechanism can be manipulated in different manners and that the function can be canceled and reactivated by counter-mutations in XXXXX (REFFFFF). Futher it has been shown that the specificity of the N-proteins can be changed from lambda to P22 by only a few mutations showing a possible coevolution, or possible interactions to increase possible genomic randomization (REFFF). (Complexicity)
Synthetic promoter library (SPL)
How does it work, examples, what have it been used to characterize? how do you construct it? Figures and illustrations to explain. Figures to explain our use? And example on our specific design primer sequences illustration on the double stranded DNA, with BB - prefix suffix.
Design and engineering of bi[o]stable
The THEORETICAL overall aim and vision
(THE bigger picture and STORY)
APPLICATIONS
how we designed our switch - selection of parts and parameters - and last presentation of our system.
Applications Unsertainties and potential problems: By designing the switch we did not know the exact distance needed from the nut-site to the terminator steam loop for proper function of anti-termination. We have taken the sequence form the natural seting and made it small enough to give sense as a biological building block.
We have tried to utilize the phage regulation to construct a biological switch that can be used in biological engineering. When considering how to characterize the subparts of our system we looked at the work already done to characterize terminator efficiency. The screening plasmids made by (REFF) endy and XXX and XXX. The work clearly demonstrates the problem by creating a weldefined data sheet system, the data achived in terms of terminator efficiency is not consistent, and shows the complexity of biology. It has not been possible for us do all the test needed to develop a wel defined switch system. Below is outlined our approach and in the end we suggest other approaches and possibilities for further work, and considerations in relation to this.
Presentation
Figures and illustrations - explanations
Designing the switch - selecting parts
(modeling) Requirements before a biological switch functions. On the paper and theoretically. Components of the switch We have decided not to use cI, why? The hong kong paper flaws!! (REF) we did not use UV-activation why? To have a stable system we did not what to use cI and UV-regulatory systems as they can impose problems with the genetic stability.
Repressor anti-Repressor system
Selecting N protein and nut site
In the end, after evaluating what component pair to use we selected λ N-protein and nut-site. Different nut-sites N-protein systems have been identified and investigated (REFFF), the nutsites for λ-phage and p21, p22, are the best described (REFFF) comparison of the antiterminator effect have not been charfully investigated, as emphasis have been on function and interacting parts. we wanted to selected the nutsite with a strong consistent anti-terminator effect. But as this was not well defined continues work was done with the lambda nut side because more articles and knowledge was available, for potential trouble shooting and improvement of the system interaction and dynamic. What have been described is that the N-nut-site pair have specific function and thus the λ-N-protein was used for continues, construction of the switch.
Flourescene protein
Why were the shown proteins selected.
The Biolector
selected filters in releation to flourescence protein
The filters for the biolector is ordered individually to fit the required needs and proteins. we decided to order the filters we needed for this application.
Filters applied for this experimentWe chose RED: Green:
Characterizing BBricks as parts of the switch
(Materials and Methods) section
Mainly the hard control of the switch is due to a double regulation system build on a both terminator-anti-terminator and repressor anti-repressor regulation. It was out of the scope of this project to construct the entire theoretical developed switch, and characterize the fully constructed switch. Have focused on characterizing the two regulatory systems individually. This was done in order to investigate if the responses were satisfactory to use in a future complete switch construction. (???? By getting the regulatory mechanism of the subparts we further, by modeling, could conclude constraints for successful function of the system and other subparts. In this section we describe the design of and the experimental setup used to characterize the subparts of the system and our bio-bricks.
Anti-terminator function
(experimental work)
selecting supparts
Why were these supparts chosen ?
AIM
Design and experimental setup
presentation - Figure of setup and explanation
Materials and methods
HOW ? what plasmids and why, what measurering method and why? refer to the notebook page with protocols - and actual info from lab.
mRNA-stability when introducing non-coding sequences problems will acour with rna-degredation of RNAP is not attracted to the area, to fast degredation, unwanted steam loops. (Reference to the terminator screening plasmids for BB)
Synthetic promoter library (SPL)
How does it work, examples, what have it been used to characterize?
how do you construct it? Figures and illustrations to explain.
Figures to explain our use? And example on our specific design primer sequences illustration on the double stranded DNA, with BB - prefix suffix.
Results
comments to the results and reference to the BB pages with info and results.
Repressor function
(experimental work)
selecting supparts
Why were these supparts chosen ?
AIM
Design and experimental setup
presentation - Figure of setup and explanation
Materials and methods
HOW ? what plasmids and why, what measurering method and why? refer to the notebook page with protocols - and actual info from lab.
Results
comments to the results and reference to the BB pages with info and results.
References
- (Gottesman et.al. 2002) Gottesman. Max E, Nudler. Evgeny, 2002 ”Transcription termination and anti-termination in E.coli” Genes to cells. (a good introduction review to termination function)
- (Franklin et.al. 1989) NC Franklin, JH Doelling - Am Soc Microbiol "Overexpression of N antitermination proteins of bacteriophages lambda, 21, and P22: loss of N protein specificity." - Journal of bacteriology, 1989
- (Jensen 2004) Ole Nørregaard Jensen, “Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry,” Current Opinion in Chemical Biology 8, no. 1 (February 2004): 33-41.
- http://syntheticbiology.org/FAQ.html
- http://www.nature.com.globalproxy.cvt.dk/nrg/journal/v6/n7/execsumm/nrg1637.html
- http://www.nature.com.globalproxy.cvt.dk/msb/journal/v2/n1/full/msb4100073.html
 "
"