Team:SDU-Denmark/labnotes8
From 2010.igem.org
(→Miniprep of "wrong" FlhDCmut) |
(→Lab notes (8/30 - 9/5)) |
||
| Line 2: | Line 2: | ||
__TOC__ | __TOC__ | ||
| + | |||
| + | == Flagella == | ||
| + | <br> | ||
| + | Since the previous FlhDCmut was wrongly mutated due to incorrect mutation primers, we are now back to square one with new correct primers. The next weeks the flagella-group are working according to the following plan: <br> | ||
| + | 1) Miniprep of plasmids with "wrong" FlhDCmut <br><br> | ||
| + | 2) Two-step PCR to get mutatet FlhDC<br> | ||
| + | 3) Cut and Ligate into pSB1C3 and pSB1AK3 and transform into TOP10 cells <br> | ||
| + | 4) Send to sequencing <br> | ||
| + | 5) Characterize biobrick <br><br> | ||
| + | === Miniprep of "wrong" FlhDCmut === | ||
| + | <br> | ||
| + | ''Done by:'' Louise <br> | ||
| + | ''Date:'' September 3rd <br> | ||
| + | ''Protocol:'' [[https://2010.igem.org/Team:SDU-Denmark/protocols#MP1.2 MP1.2]] <br> | ||
| + | ''Notes:'' <br> | ||
| + | No changes of protocol were made. <br> | ||
| + | ''Results:'' | ||
| + | Nanodrop after sample was dried down: <br> | ||
| + | '''Sample 1:''' Concentration: 192ng/ul Purity: 1.96/1.90 <br> | ||
| + | '''Sample 2:''' Concentration: 200ng/ul Purity: 1.84/1.88 <br><br> | ||
| + | The samples were run on a 1.5% gel with a 10kb ladder. The bands are positioned between 1500bp and 1200bp. FlhDCmut in pSB1C3 is 1248bp. <br> | ||
| + | [[Image:Team SDU-Denmark Miniprep af flhDCmut i 1C3.jpg|300px]] | ||
| + | ---- | ||
| + | |||
| + | ==== Two-step PCR of miniprep ==== | ||
| + | <br> | ||
| + | ===== First step: PCR of miniprep with mutation primers ===== | ||
| + | <br> | ||
| + | ''Done by:'' Louise <br> | ||
| + | ''Date:'' September 7th <br> | ||
| + | ''Protocol:'' [[https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.1 CP1.1]] <br> | ||
| + | ''Notes:'' <br> | ||
| + | '''3 x Premix 1:''' <br> | ||
| + | 114.3ul water <br> | ||
| + | 15ul Pfu Buffer <br> | ||
| + | 4.5ul dNTP <br> | ||
| + | 3ul MgSO4 <br> | ||
| + | 4.5ul FlhDC fw <br> | ||
| + | 4.5ul FlhDCmut rev <br> | ||
| + | 1.2ul PFU <br> | ||
| + | 3ul template (miniprep) <br><br> | ||
| + | '''3 x Premix 2:''' <br> | ||
| + | 114.3ul water <br> | ||
| + | 15ul Pfu Buffer <br> | ||
| + | 4.5ul dNTP <br> | ||
| + | 3ul MgSO4 <br> | ||
| + | 4.5ul FlhDCmut fw <br> | ||
| + | 4.5ul FlhDC rev <br> | ||
| + | 1.2ul PFU <br> | ||
| + | 3ul template (miniprep) <br><br> | ||
| + | ''Results:'' | ||
| + | <br> | ||
| + | The PCR samples were run on a 1.5% gel with a 100bp-1000bp ladder. All samples looked okay and were pooled as sample 1.1 and sample 2.1 <br> | ||
| + | NanoDrop: <br> | ||
| + | '''Sample 1.1:''' Concentration: 359.5ng/ul Purity: 1.83/2.20 <br> | ||
| + | '''Sample 2.1:''' Concentration: 304.1ng/ul Purity: 1.85/2.23 <br> | ||
| + | |||
| + | ===== Second step: 2-step PCR with mutated template sample 1.1 and 2.1 and FlhDC fw and rev primers ===== | ||
| + | <br> | ||
| + | ''Done by:'' Maria <br> | ||
| + | ''Date:'' September 9th <br> | ||
| + | ''Protocol:'' [[https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.1 CP1.1]] <br> | ||
| + | ''Notes:'' <br> | ||
| + | '''Premix:''' <br> | ||
| + | 38ul water <br> | ||
| + | 5ul PFU buffer + MgSO4 <br> | ||
| + | 1.5ul dNTP <br> | ||
| + | 1ul Sample 1.1 <br> | ||
| + | 1ul Sample 2.1 <br> | ||
| + | 1.5ul FlhDC fw primer <br> | ||
| + | 1.5ul FlhDC rev primer <br> | ||
| + | 0.5ul PFU <br><br> | ||
| + | '''PCR Program:''' <br> | ||
| + | <table style="text-align: left; height: 260px; width: 225px;" border="1" | ||
| + | cellpadding="2" cellspacing="2"> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">1:Start<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">95C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;">2 min<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">2: Denaturing<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">95C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;">30 sec<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">3: Annealing<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">56C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;">30 sec<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">4: Elongation<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">72C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;">2 min<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">5:<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">GO TO<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;">2 rep. 4x<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">6: Denaturing<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">95C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;">30 sec<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">7: Annealing<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">63C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;">30 sec<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">8: Elongation<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">72C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;">2 min<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">9:<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">GO TO<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;">6 rep. 25x<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">10: End <br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">72C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;">5 min<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">12: Hold<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">4C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;"><br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> <br><br> | ||
| + | |||
| + | ''Results:'' <br> | ||
| + | Some of the PCR product was run on a 1.5% gel with a 10kb ladder. <br> | ||
| + | The rest of the product was extracted from a new gel. | ||
| + | |||
| + | ===== PCR of Gel extraction ===== | ||
| + | <br> | ||
| + | ''Done by:'' Maria <br> | ||
| + | ''Date:'' September 9th <br> | ||
| + | ''Protocol:'' [[https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.1 CP1.1]] <br> | ||
| + | ''Notes:'' <br> | ||
| + | '''Premix x 6:''' <br> | ||
| + | 234ul water <br> | ||
| + | 30ul PFU buffer + MgSO4 <br> | ||
| + | 9ul dNTP <br> | ||
| + | 9ul FlhDC fw primer <br> | ||
| + | 9ul FlhDC rev primer <br> | ||
| + | 2.5ul PFU <br> | ||
| + | 6ul template <br> | ||
| + | |||
| + | <table style="text-align: left; height: 260px; width: 225px;" border="1" | ||
| + | cellpadding="2" cellspacing="2"> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">1:Start<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">95C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;">2 min<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">2: Denaturing<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">95C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;">30sec<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">3: Annealing<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">63C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;">30 sec<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">4: Elongation<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">72C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;">2 min<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">5:<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">GO TO<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;">2 rep. 29x<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">6: End <br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">72C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;">5 min<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 102px;">7: Hold<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">4C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 45px;"><br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> <br><br> | ||
| + | <br> | ||
== Photosensor == | == Photosensor == | ||
=== PCR on pKJ606 with PSfw and VR primers === | === PCR on pKJ606 with PSfw and VR primers === | ||
| Line 246: | Line 490: | ||
<br> | <br> | ||
Results: Primers were confirmed working, the next step will be PFU pcr.<br> | Results: Primers were confirmed working, the next step will be PFU pcr.<br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Revision as of 09:03, 1 October 2010
Lab notes (8/30 - 9/5)
Contents |
Flagella
Since the previous FlhDCmut was wrongly mutated due to incorrect mutation primers, we are now back to square one with new correct primers. The next weeks the flagella-group are working according to the following plan:
1) Miniprep of plasmids with "wrong" FlhDCmut
2) Two-step PCR to get mutatet FlhDC
3) Cut and Ligate into pSB1C3 and pSB1AK3 and transform into TOP10 cells
4) Send to sequencing
5) Characterize biobrick
Miniprep of "wrong" FlhDCmut
Done by: Louise
Date: September 3rd
Protocol: [MP1.2]
Notes:
No changes of protocol were made.
Results:
Nanodrop after sample was dried down:
Sample 1: Concentration: 192ng/ul Purity: 1.96/1.90
Sample 2: Concentration: 200ng/ul Purity: 1.84/1.88
The samples were run on a 1.5% gel with a 10kb ladder. The bands are positioned between 1500bp and 1200bp. FlhDCmut in pSB1C3 is 1248bp.
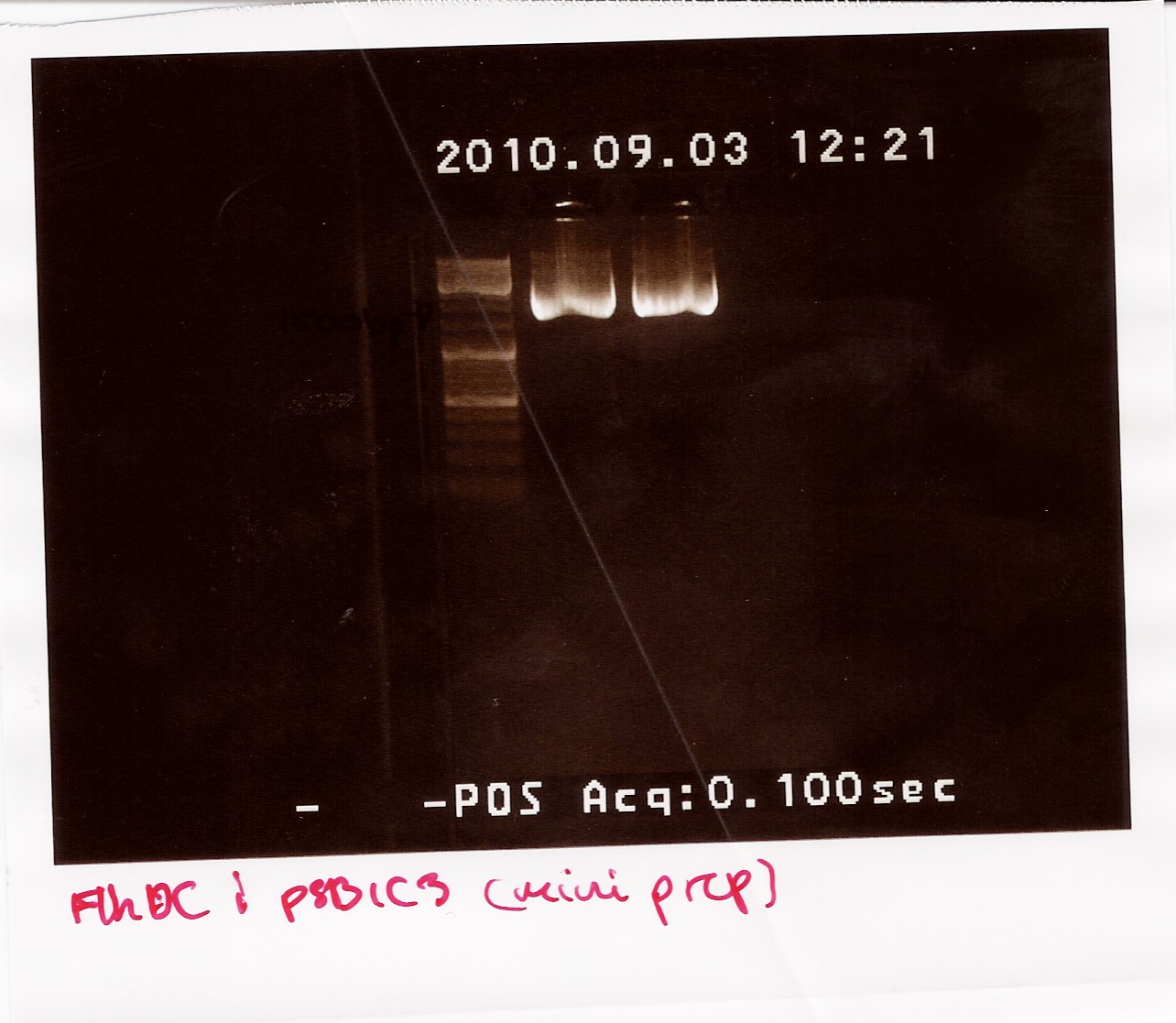
Two-step PCR of miniprep
First step: PCR of miniprep with mutation primers
Done by: Louise
Date: September 7th
Protocol: [CP1.1]
Notes:
3 x Premix 1:
114.3ul water
15ul Pfu Buffer
4.5ul dNTP
3ul MgSO4
4.5ul FlhDC fw
4.5ul FlhDCmut rev
1.2ul PFU
3ul template (miniprep)
3 x Premix 2:
114.3ul water
15ul Pfu Buffer
4.5ul dNTP
3ul MgSO4
4.5ul FlhDCmut fw
4.5ul FlhDC rev
1.2ul PFU
3ul template (miniprep)
Results:
The PCR samples were run on a 1.5% gel with a 100bp-1000bp ladder. All samples looked okay and were pooled as sample 1.1 and sample 2.1
NanoDrop:
Sample 1.1: Concentration: 359.5ng/ul Purity: 1.83/2.20
Sample 2.1: Concentration: 304.1ng/ul Purity: 1.85/2.23
Second step: 2-step PCR with mutated template sample 1.1 and 2.1 and FlhDC fw and rev primers
Done by: Maria
Date: September 9th
Protocol: [CP1.1]
Notes:
Premix:
38ul water
5ul PFU buffer + MgSO4
1.5ul dNTP
1ul Sample 1.1
1ul Sample 2.1
1.5ul FlhDC fw primer
1.5ul FlhDC rev primer
0.5ul PFU
PCR Program:
| 1:Start |
95C |
2 min |
| 2: Denaturing |
95C |
30 sec |
| 3: Annealing |
56C |
30 sec |
| 4: Elongation |
72C |
2 min |
| 5: |
GO TO |
2 rep. 4x |
| 6: Denaturing |
95C |
30 sec |
| 7: Annealing |
63C |
30 sec |
| 8: Elongation |
72C |
2 min |
| 9: |
GO TO |
6 rep. 25x |
| 10: End |
72C |
5 min |
| 12: Hold |
4C |
Results:
Some of the PCR product was run on a 1.5% gel with a 10kb ladder.
The rest of the product was extracted from a new gel.
PCR of Gel extraction
Done by: Maria
Date: September 9th
Protocol: [CP1.1]
Notes:
Premix x 6:
234ul water
30ul PFU buffer + MgSO4
9ul dNTP
9ul FlhDC fw primer
9ul FlhDC rev primer
2.5ul PFU
6ul template
| 1:Start |
95C |
2 min |
| 2: Denaturing |
95C |
30sec |
| 3: Annealing |
63C |
30 sec |
| 4: Elongation |
72C |
2 min |
| 5: |
GO TO |
2 rep. 29x |
| 6: End |
72C |
5 min |
| 7: Hold |
4C |
Photosensor
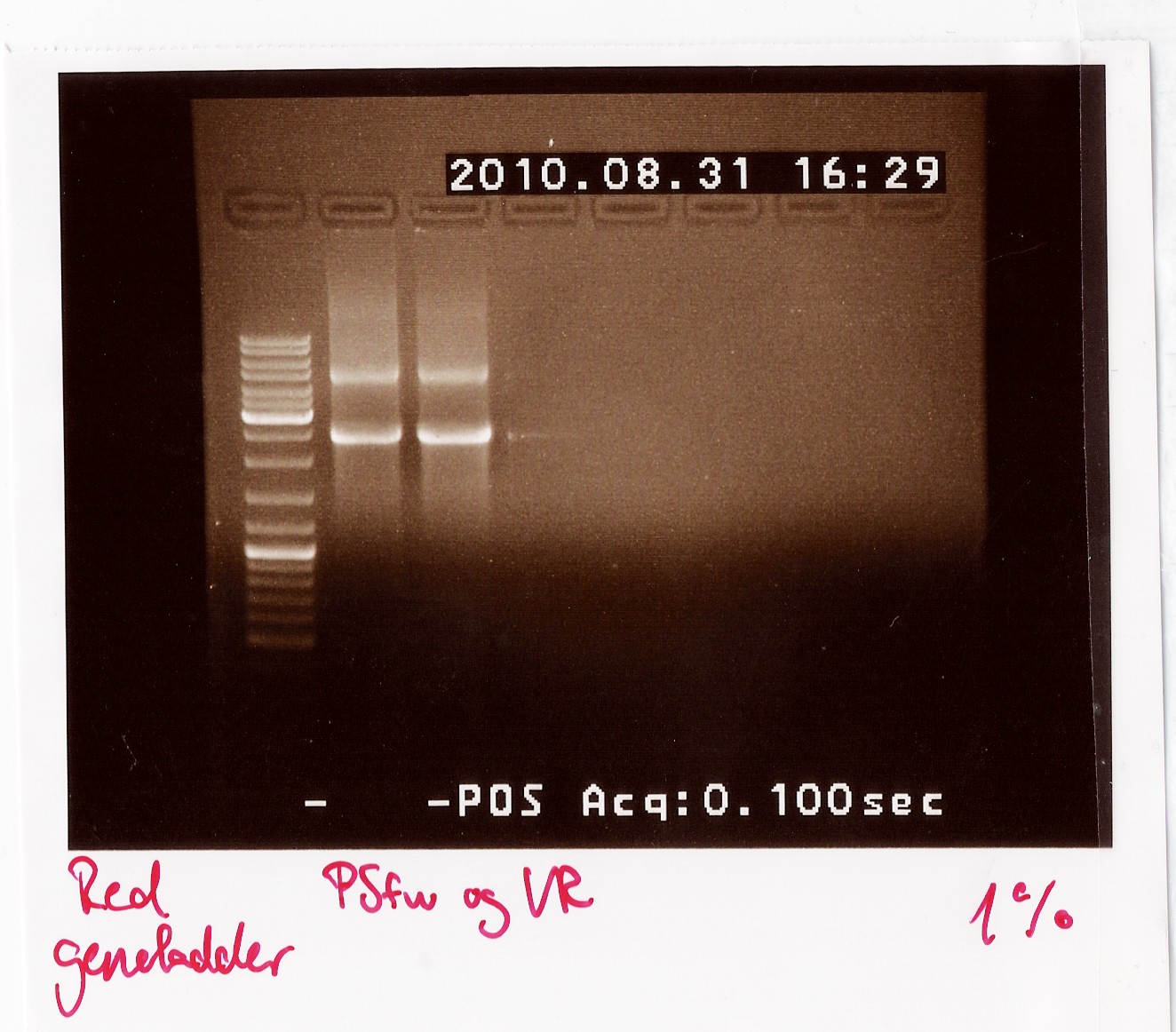
PCR on pKJ606 with PSfw and VR primers
Date: 31/8
https://2010.igem.org/wiki/index.php?title=Team:SDU-Denmark/labnotes8&action=edit§ion=3
Done by: LC
Methods: PCR
Protocols: CP1.3[1]
Notes:
Premix:
7,5 µl 10xTAQ Buffer
3 µl MgCl2
3 µl VF2
3 µl VR
1,5 µl dNTP
55,5 µl H2O
3 µl template (PS1 miniprep)
3/8 µl TAQ Polymerase
PCR Program:
| Start |
94 C |
2 min |
| Denaturing |
94 C |
1 min |
| Annealing |
55 C |
1 min |
| Elongation |
72 C |
3 min |
| Goto2 |
rep |
29x |
| End |
72 C |
3 min |
| Hold |
4 C |
Results: The bands that showed up were around 2500 BP as expected from the sequencing results. This means that the designed primers have a high possibility of working, so that they will be ordered.
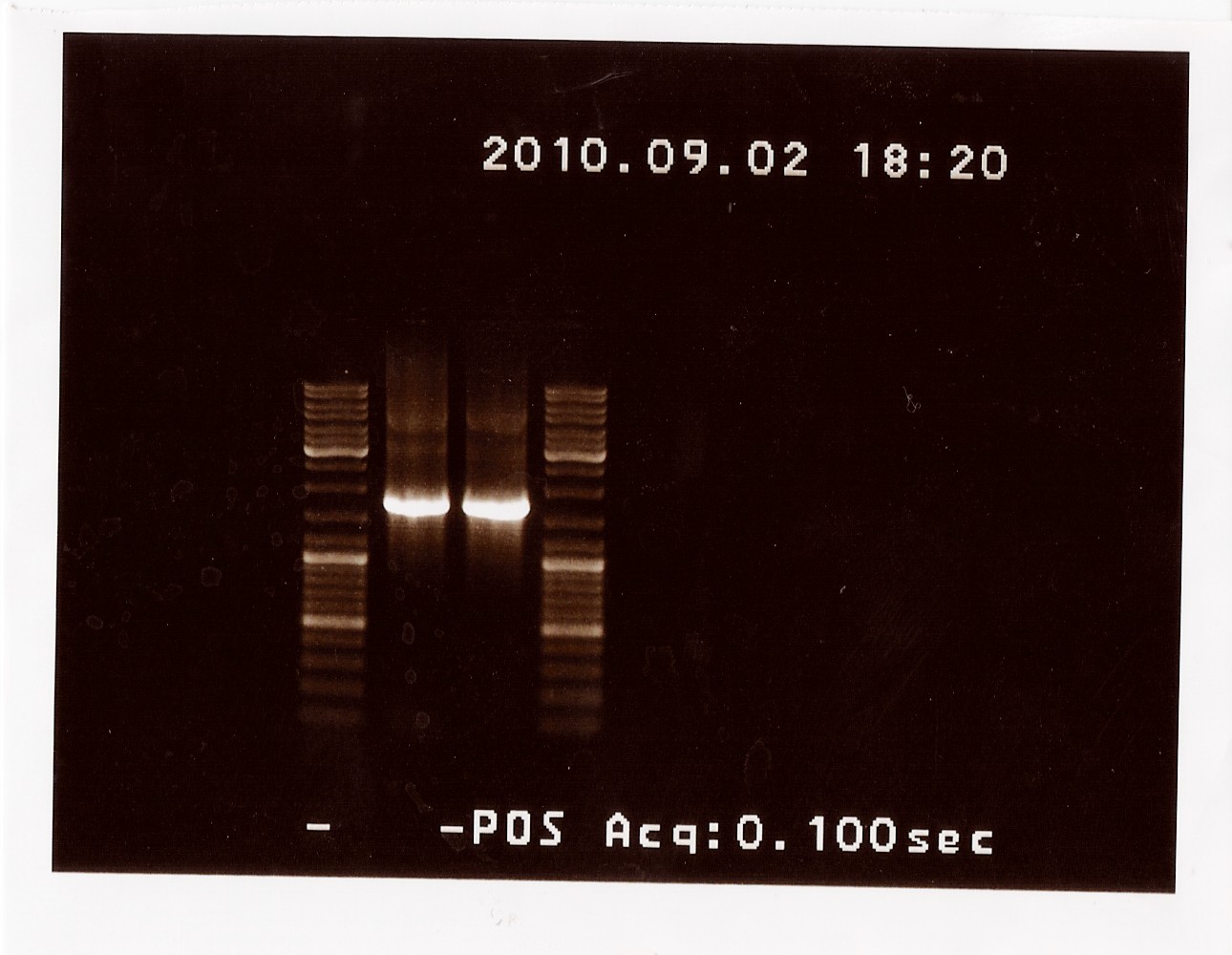
PCR on pKJ606 with PSfw and PSrv primers
Date: 02/09
Done by: LC
Methods: PCR
Protocols: CP1.3[2]
Notes:
Premix:
7,5 µl 10xTAQ Buffer
3 µl MgCl2
3 µl VF2
3 µl VR
1,5 µl dNTP
55,5 µl H2O
3 µl template (PS1 miniprep)
3/8 µl TAQ Polymerase
PCR Program:
| Start |
94 C |
2 min |
| Denaturing |
94 C |
1 min |
| Annealing |
55 C |
1 min |
| Elongation |
72 C |
2 min |
| Goto2 |
rep |
29x |
| End |
72 C |
3 min |
| Hold |
4 C |
Results: The bands that showed up were around 1750 BP, further confirming sequencing results. New primers for taking the whole gene out have been ordered.
PCR on pKJ606 with fwPS2 and rvPS2 primers
Date: 04/09
Done by: LC
Methods: PCR
Protocols: CP1.3[3]
Notes:
Premix:
12,5 µl 10xTAQ Buffer
5 µl MgCl2
5 µl VF2
5 µl VR
2,5 µl dNTP
59,5 µl H2O
4 µl template (Consisting of 2 µl H2O and 2 µl of pKJ606 miniprep)
1/2 µl TAQ Polymerase
PCR Program:
| Start |
94 C |
2 min |
| Denaturing |
94 C |
1 min |
| Annealing |
55 C |
1 min |
| Elongation |
72 C |
2:30 min |
| Goto2 |
rep |
29x |
| End |
72 C |
3 min |
| Hold |
4 C |
Results: Primers were confirmed working, the next step will be PFU pcr.
 "
"