Team:ETHZ Basel/Modeling/Chemotaxis
From 2010.igem.org
(→Modeling of the chemotactic receptor pathway) |
|||
| Line 5: | Line 5: | ||
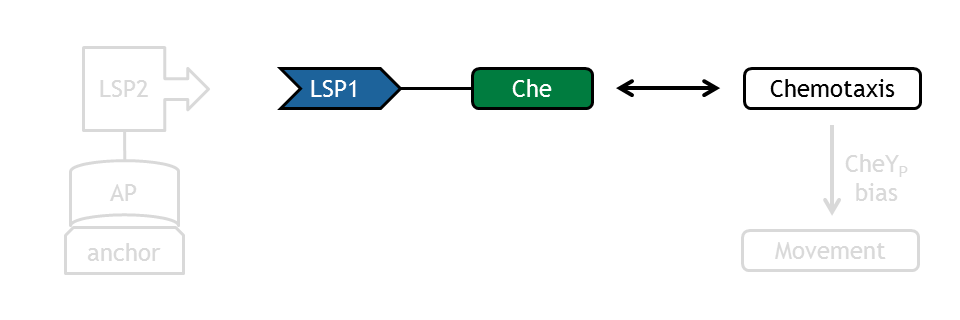
[[Image:ETHZ_Basel_molecular_che.png|thumb|400px|'''Schematical overview of the light switch device and the interaction with the chemotaxis pathway.''' LSP refers to light switch protein, AP to anchor protein and Che to the attacked protein of the chemotaxis pathway.]] | [[Image:ETHZ_Basel_molecular_che.png|thumb|400px|'''Schematical overview of the light switch device and the interaction with the chemotaxis pathway.''' LSP refers to light switch protein, AP to anchor protein and Che to the attacked protein of the chemotaxis pathway.]] | ||
| - | The chemotactic receptor pathway in E. coli is quite complex. Published models of chemotaxis [[Team:ETHZ Basel/Modeling/Chemotaxis#References|[1]]], [[Team:ETHZ Basel/Modeling/Chemotaxis#References|[2]]], [[Team:ETHZ Basel/Modeling/Chemotaxis#References|[3]]], [[Team:ETHZ Basel/Modeling/Chemotaxis#References|[4]]] thus have to | + | The chemotactic receptor pathway in E. coli is quite complex. Published models of chemotaxis [[Team:ETHZ Basel/Modeling/Chemotaxis#References|[1]]], [[Team:ETHZ Basel/Modeling/Chemotaxis#References|[2]]], [[Team:ETHZ Basel/Modeling/Chemotaxis#References|[3]]], [[Team:ETHZ Basel/Modeling/Chemotaxis#References|[4]]] thus have to do many assumptions in order to suit the question of the investigation. It was therefore decided to implement four different models to be able to achieve a more general consensus prediction of chemotactic behavior in E. lemming. |
For the combined model implementation, only two chemotaxis models have been further investigated: Spiro et al. (1997) and Mello & Tu (2003). | For the combined model implementation, only two chemotaxis models have been further investigated: Spiro et al. (1997) and Mello & Tu (2003). | ||
| Line 13: | Line 13: | ||
|- valign="top" | |- valign="top" | ||
|[[Image:ETHZ_Basel_chemotaxis_spiro_zero_top.png|thumb|260px|'''Response of the system with no aspartate present.''' Removal of a species has a significant effect on the chemotactic receptor pathway.]] | |[[Image:ETHZ_Basel_chemotaxis_spiro_zero_top.png|thumb|260px|'''Response of the system with no aspartate present.''' Removal of a species has a significant effect on the chemotactic receptor pathway.]] | ||
| - | |[[Image:ETHZ_Basel_chemotaxis_spiro_small_top.png|thumb|260px|'''Response of the system with a small aspartate step.''']] | + | |[[Image:ETHZ_Basel_chemotaxis_spiro_small_top.png|thumb|260px|'''Response of the system with a small aspartate step.''' Removal of a species has a lower effect than without presence of aspartate.]] |
| - | |[[Image:ETHZ_Basel_chemotaxis_spiro_high_top.png|thumb|260px|'''Response of the system with a big aspartate step.''']] | + | |[[Image:ETHZ_Basel_chemotaxis_spiro_high_top.png|thumb|260px|'''Response of the system with a big aspartate step.''' Saturation or near-saturation of the aspartate receptor changes the effect of the removal of a species significantly.]] |
|- | |- | ||
|[[Image:ETHZ_Basel_chemotaxis_spiro_zero_down.png|thumb|260px|'''Receptor proteins of the system with no aspartate present.''']] | |[[Image:ETHZ_Basel_chemotaxis_spiro_zero_down.png|thumb|260px|'''Receptor proteins of the system with no aspartate present.''']] | ||
Revision as of 07:34, 29 September 2010
Modeling of the chemotactic receptor pathway
The chemotactic receptor pathway in E. coli is quite complex. Published models of chemotaxis [1], [2], [3], [4] thus have to do many assumptions in order to suit the question of the investigation. It was therefore decided to implement four different models to be able to achieve a more general consensus prediction of chemotactic behavior in E. lemming.
For the combined model implementation, only two chemotaxis models have been further investigated: Spiro et al. (1997) and Mello & Tu (2003).
Spiro et al. (1997) [1]
The model from Spiro et al. (1997) [1] has been adapted to identify candidates of the chemotactic receptor pathway by enabling removal of a species upon light-induction. It basically predicts that all Che proteins (CheR, B, Y and Z) would be a suitable candidate. For CheR and CheY, the concentration of CheYp drops more than Delta (initial value - threshold), for CheB and CheZ it increases more than Delta. For all Che proteins, the concentrations stay below/above the threshold, until they are deactivated with far-red light. The best results were obtained for assuming a high ligand concentration (saturation, the methylation level of the receptors is high). For CheY and CheZ the reaction times were much faster than for CheB and CheR.
Mello & Tu (2003) [2]
File:ETHZ Basel chemotaxis mello small top.png Response of the system with medium aspartate levels. | File:ETHZ Basel chemotaxis mello high top.png Response of the system with high aspartate levels. | |
File:ETHZ Basel chemotaxis mello small down.png Receptor proteins of the system with medium aspartate levels. | File:ETHZ Basel chemotaxis mello high down.png Receptor proteins of the system with high aspartate levels. |
Rao et al. (2004) [3]
This model predicts quite a similar behavior of the Che proteins, as demonstrated by the previous model. A notable difference is that the response of CheYp level to the change of the level of the other Che proteins is robust to the fluctuations of the ligand (attractant) concentration. In the previous model (Spiro), the most desirable results were obtained only for higher concentrations of the ligand.
Barkai & Leibler (1997) [4]
The model is implemented, but not yet documented.
References
[1] [http://www.pnas.org/content/94/14/7263.full Spiro et al: A model of excitation and adaptation in bacterial chemotaxis. PNAS 1997 94;14;7263-7268.]
[2] [http://www.cell.com/biophysj/retrieve/pii/S0006349503700216 Mello & Tu: Perfect and Near-Perfect Adaptation in a Model of Bacterial Chemotaxis. Biophysical Journal 2003 84;5;2943-2956.]
[3] [http://www.plosbiology.org/article/info:doi/10.1371/journal.pbio.0020049 Rao et al: Design and Diversity in Bacterial Chemotaxis. PLoS Biol 2004;2;2;239-252.]
[4] [http://www.nature.com/nature/journal/v387/n6636/abs/387913a0.html Barkai & Leibler: Robustness in simple biochemical networks. Nature 1997;387;913-917.]
 "
"