Team:ETHZ Basel/Modeling
From 2010.igem.org
| Line 8: | Line 8: | ||
[[Image:Modeling_overview_molecular.png|thumb|400px|'''Schematical overview of the devices and change upon light pulse induction.''']] | [[Image:Modeling_overview_molecular.png|thumb|400px|'''Schematical overview of the devices and change upon light pulse induction.''']] | ||
| - | The core component of E. lemming is the fusion of one light-sensitive protein (LSP?) to a protein of the chemotaxis pathway (Che?). Upon change of wavelength of light pulses, this component will dimerize with the corresponding light-sensitive protein (LSP?'), which is linked to an anchor protein, bound to an anchor (plasmid). The result is a change of the spatial localization of Che? and perturbation of the chemotaxis pathway, which ultimately leads to a different tumbling/directed flagellar movement ratio. | + | The core component of E. lemming is the fusion of one light-sensitive protein (LSP?) to a protein of the chemotaxis pathway (Che?). Upon change of wavelength of light pulses, this component will dimerize with the corresponding light-sensitive protein (LSP?'), which is linked to an anchor protein, bound to an anchor (plasmid). The result is a change of the spatial localization of Che? and perturbation of the chemotaxis pathway, which ultimately leads to a different tumbling/directed flagellar movement state ratio. |
| + | |||
| + | In a first step, we implemented individual deterministic molecular models of subdevices. | ||
| + | * [[Team:ETHZ_Basel/Modeling/Light_switch|'''Light switch''']]: based upon the light-sensitive dimerizing Arabidopsis proteins PhyB and PIF3. | ||
| + | * [[Team:ETHZ_Basel/Modeling/Chemotaxis|'''Chemotaxis''']]: | ||
| + | * [[Team:ETHZ_Basel/Modeling/Movement|'''Movement''']]: | ||
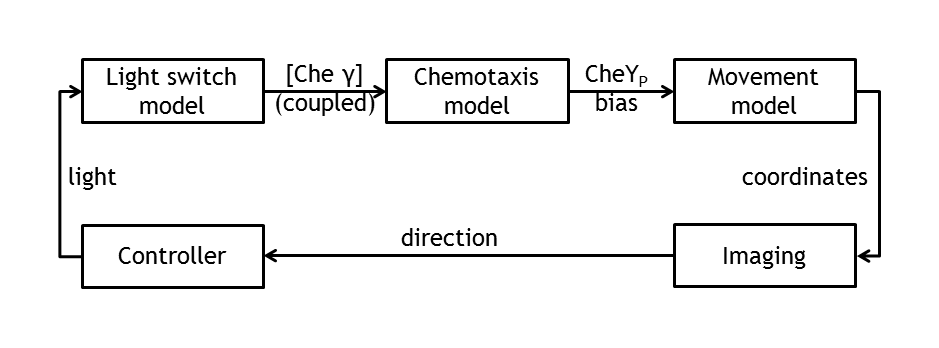
[[Image:Modeling_overview.png|thumb|400px|'''Combined models.''' Coupled individual models for the simulation of the whole process and their interfaces.]] | [[Image:Modeling_overview.png|thumb|400px|'''Combined models.''' Coupled individual models for the simulation of the whole process and their interfaces.]] | ||
Revision as of 12:32, 22 September 2010
Molecular Modeling Overview
In order to support wet laboratory experiments and to create a test bench for the information processing part, a molecular model of E. lemming was created. This goal was achieved by implementing and combining deterministic molecular models of the individual parts.
Implementation of molecular models
The core component of E. lemming is the fusion of one light-sensitive protein (LSP?) to a protein of the chemotaxis pathway (Che?). Upon change of wavelength of light pulses, this component will dimerize with the corresponding light-sensitive protein (LSP?'), which is linked to an anchor protein, bound to an anchor (plasmid). The result is a change of the spatial localization of Che? and perturbation of the chemotaxis pathway, which ultimately leads to a different tumbling/directed flagellar movement state ratio.
In a first step, we implemented individual deterministic molecular models of subdevices.
- Light switch: based upon the light-sensitive dimerizing Arabidopsis proteins PhyB and PIF3.
- Chemotaxis:
- Movement:
In the theory world, the steps we are following in mindlessly driving E.coli to our pre - defined target are the following:
• deterministic (ODE) & stochastic models of the chemotaxis pathway (documented from the literature)
• model of the movement of E.coli (built on the information of the pathway derived from the molecular models)
• control algorithms ( built on the user’s desire to play around with E.coli)
• image tracking & image processing algorithms
• java applications/movies of the E.Lemming (the fun part)
 "
"