Team:Stockholm/21 August 2010
From 2010.igem.org
(→Gel verification) |
(→Gel verification) |
||
| Line 33: | Line 33: | ||
=====Gel verification===== | =====Gel verification===== | ||
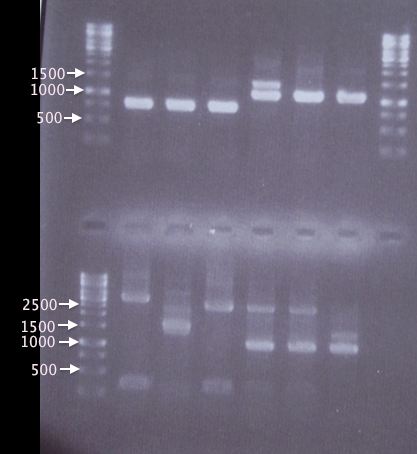
| - | [[image:ColPCR_Hisass_19aug.png|200px|thumb|right|''' | + | [[image:ColPCR_Hisass_19aug.png|200px|thumb|right|'''Gel verification of SOD*His, yCCS*His, His*SOD and His*yCCS assemblies into pSB1K3 vector'''<br />'''1st row:'''λ, A1, A2, A3, A4, A5, A6, λ.<br />'''2nd row:'''λ, A7, A8, A9, A10, A11, A12, A13.<br />3 μl λ, 5 μl sample.<br />λ=O'GeneRuler 1 kb DNA ladder.]] |
====Transfer of RFP to pEX==== | ====Transfer of RFP to pEX==== | ||
Revision as of 17:20, 22 August 2010
Contents |
Hassan
found some extra articles for gene-regulatory networks: [http://www.ncbi.nlm.nih.gov/pubmed/17504165] and [http://www.biomedcentral.com/1471-2105/8/S6/S9]
Andreas
Assembly of CPP⋅protein⋅His constructs
Assembly of His⋅SOD/yCCS and SOD/yCCS⋅His into pSB1K3
Step I in cloning strategy (19/8)
Transformation results
- pSB1K3.SOD⋅His
- pSB1K3.yCCS⋅His
- pSB1K3.His⋅SOD
- pSB1K3.His⋅yCCS
Good yield on all plates.
Colony PCR
Three white colonies picked from each plate for verification.
- pSB1K3.SOD⋅His: 1=A1, 2=A2, 3=A3
- pSB1K3.yCCS⋅His: 1=A4, 2=A5, 3=A6
- pSB1K3.His⋅SOD: 1=A7, 2=A8, 3=A9
- pSB1K3.His⋅yCCS: 1=A10, 2=A11, 3=A12
- Blank: A13
Procedures as described in colony PCR protocol.
- Forward primer: VF2
- Reverse primer: VR
- Elongation time: 2:00
Gel verification
Transfer of RFP to pEX
Continued from 19/8
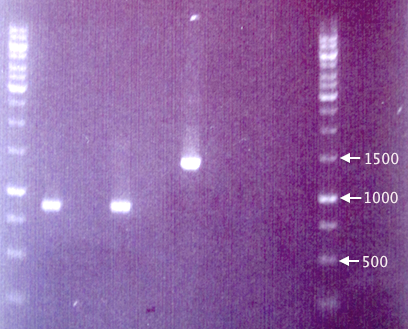
Colony PCR
Since the gel results from 20/8 only verified pMA.His, a new colony PCR was run to verify pEX and pEX.RFP. Two new clones (5 & 6) of pEX.RFP were picked and run with clones 2 and 3 from 20/8. To test whether the red colonies could possibly be from pSB1K3 (i.e. if the selection didn't work properly), pEX.RFP 2 was also amplified with pSB primer VF2 and VR.
Also, pEX plasmid was amplified in parallel with pEX plasmid to compare the insert sizes.
- J1: pEX.RFP 2
- J8: pEX.RFP 3
- J2: pEX.RFP 5
- J3: pEX.RFP 6
- J4: pEX clone
- J5: pEX plasmid
- J6: pEX.RFP 2 (w/ pSB primers)
- J7: Blank
Procedures as described in colony PCR protocol:
- Forward primer: pEXf or pSB VF2
- Reverse primer: pEXr or pSB VR
- Elongation: 2:00
Gel verification
1 % agarose, 90 V, 1 h.
Expected bands:
- pEX.RFP: 1272 bp
- pEX (w/ insert): 1414 bp
Results:
The fragments seen for samples J1 and J2 are considerably shorter (≈850 bp) than the expected 1272 bp, making it very unlikely that they contain the full-length RFP cassettes. This is strange, since the colonies picked were indeed red, obviously expressing RFP.
The results from pEX vector samples are also quite strange. The pEX clone (J4) resulted in a band corresponding well to the expected 1414 bp. However, the pEX plasmid (J5) does not result in any band whatsoever.
A new gel will be run and possibly a new transfer of RFP to pEX attempted.
Colony re-streak
In order to verify that our red colonies are indeed carrying pEX and not pSB1K3 (i.e. the Amp selection has failed), resulting in false-positives for transfer of RFP from pSB1K3 to pEX, clones 2, 3, 5 and 6 were streaked on quarters of Amp 100 and Km 50 plates. Plates incubated 48 h in 37 °C.
If RFP has been successfully transfered to pEX, the clones should only grow on Amp 100 plates.
 "
"