Team:WashU/Splicing/Project Goals
From 2010.igem.org
(New page: {{WashUHeader}} =Splicing, the New Alternative= The 2010 WashU iGEM team is attempting to create and modulate a synthetic splicing mechanism in S. cerevisiae. This will add a valuable new...)
Newer edit →
Revision as of 16:49, 16 August 2010

Splicing, the New Alternative
The 2010 WashU iGEM team is attempting to create and modulate a synthetic splicing mechanism in S. cerevisiae. This will add a valuable new tool into the synthetic biologist's tool-belt that can be utilized in many different applications.
Constructs
Three different genetic constructs are being made and tested by this years iGEM team. Each one builds upon the previous construct in our effort to develop and showcase alternative splicing as a synthetic biology tool.
Construct 1
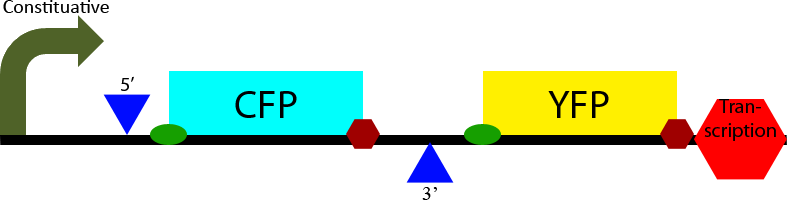
Construct 1 seeks to demonstrate that splicing of the designed intron occurs in S. cerevisiae. To accomplish this it contains two open reading frames, one which codes for CFP and one which codes for YFP. The CFP open reading frame is contained within an intron, causing it to be spliced out. In eukaryotes only the first open reading frame is translated, so in the non-spliced mRNA CFP will be translated and YFP will not be expressed. However in the spliced transcript CFP will be spliced out and YFP will be expressed. Observation of YFP will demonstrate that splicing of the designed intron has occurred within the cell. Observation of CFP will demonstrate that splicing of the designed intron is not 100% effective.
Construct 2
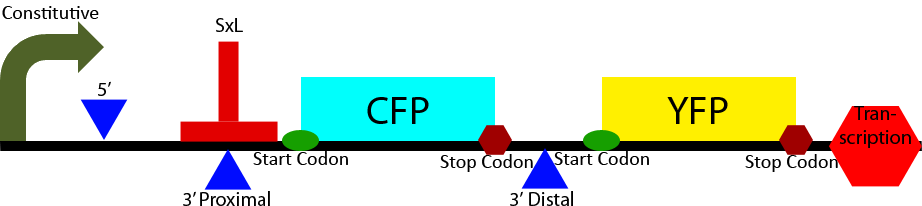
Construct 2 seeks to demonstrate the affect of Sex-Lethal (SxL) expression on 3' splice site selection. It contains a proximal and distal 3' splice sites. The proximal 3' splice site contains a SxL recognition sequence. Initially the proximal 3' splice site is favored due to its greatly decreased intron length (Klinz, Gallwitz, 1985) causing a greater CFP to YFP ratio. Upon expression of SxL the 3' proximal site will be inhibited due to competitive binding between SxL and U2AF (see Project Background section). This will cause a shift in the ratio of CFP to YFP towards YFP demonstrating synthetic controllable alternative splicing within S. cerevisiae.
Construct 3
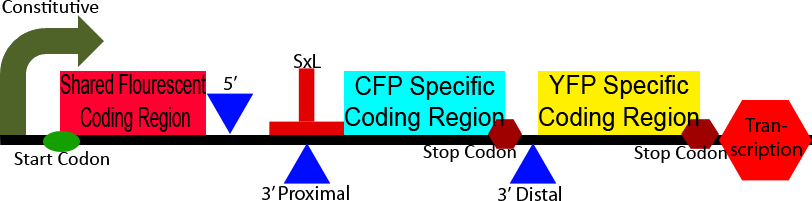
Construct 3 demonstrates an application of our alternative splicing system in order to show the unique advantages that alternative splicing can confer. This construct differs from construct 2 in the CFP and YFP proteins and split between a homologous 5' region and heterologous 3' regions. The 5' homologous region is upstream of the splice sites, causing it to always be include in the processed mRNA, the 3' ends of the two florescent proteins are then alternatively spliced on, leadings to the production of either CFP or YFP. This shows the production of two different isoforms of a fluorescent protein, the ratio of which can be modulated through the use of a splicing regulatory protein.
SxL Construct
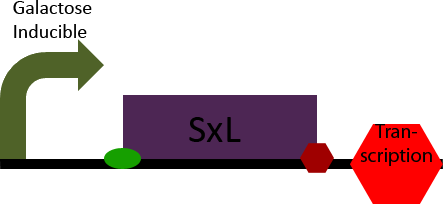
The SxL construct simply allows inducible expression of the SxL protein in S. cerevisiae. It accomplishes this by placing the D. melanogaster gene SxL under the control of a galactose inducible promoter.
Biobricks
The following biobricks were used in the above constructs:
- The Constitutive Yeast ADH1 Promoter. [http://partsregistry.org/wiki/index.php?title=Part:BBa_J63005 BBa_J63005]
- The Inducible Yeast Gal1 Promoter. [http://partsregistry.org/wiki/index.php?title=Part:BBa_J63006 BBa_J63006]
- The Designed Yeast Kozak Sequence. [http://partsregistry.org/wiki/index.php?title=Part:BBa_J63003 Part:BBa_J63003]
- Engineered Cyan Fluorescent Protein derived from A. victoria GFP. [http://partsregistry.org/wiki/index.php?title=Part:BBa_E0020 BBa_E0020]
- Enhanced Yellow Fluorescent Protein derived from A. victoria GFP. [http://partsregistry.org/wiki/index.php?title=Part:BBa_E0030 BBa_E0030]
- The Yeast ADH1 Terminator. [http://partsregistry.org/wiki/index.php?title=Part:BBa_J63002 BBa_J63002]
Works Cited
Klinz, Gallwitz, 1985. Size and position of intervening sequences are critical for the splicing efficiency of pre-mRNA in the yeast Saccharomyces cerevisiae. Nucleic acids research, v.13, 3791-3804
 "
"