Team:TU Delft/project/genetic regulation
From 2010.igem.org
| Line 21: | Line 21: | ||
The Crp-protein is a global regulator ubiquitous to ''E.coli'' which is known to bind to regions of promoters known to activate genes involved in the degradation of non-glucose carbon sources, in this way activating the genes downstream of it. We would like to seamlessly integrate the expression of our alkane degrading genes into the ''E.coli'' system by making the alkane-degrading genes sensitive to Crp. | The Crp-protein is a global regulator ubiquitous to ''E.coli'' which is known to bind to regions of promoters known to activate genes involved in the degradation of non-glucose carbon sources, in this way activating the genes downstream of it. We would like to seamlessly integrate the expression of our alkane degrading genes into the ''E.coli'' system by making the alkane-degrading genes sensitive to Crp. | ||
| + | |||
| + | |||
| + | ====Modelling==== | ||
| + | The modelling of the sensing system is described [[Team:TU_Delft/project/modeling/sensing|here]] | ||
====STEP 1: PalkS / AlkS BioBrick==== | ====STEP 1: PalkS / AlkS BioBrick==== | ||
| Line 48: | Line 52: | ||
* PalkS / AlkS: E.coli K12/310C | * PalkS / AlkS: E.coli K12/310C | ||
* PalkS / AlkS / PalkB: E.coli K12/311C | * PalkS / AlkS / PalkB: E.coli K12/311C | ||
| - | * P(CaiF) / AlkS / PalkB: | + | * P(CaiF) / AlkS / PalkB: E.coli K12/312C |
* Negative control: E.coli K12 | * Negative control: E.coli K12 | ||
The characterization process of the hydrocarbon sensing sub-project will entail output measurements of fluorescence at varying octane concentrations (0.1% – 1%) of the abovementioned strains. These measurements will be performed using a 96-well plate reader. Growth curves will be followed by OD-600 alongside the fluorescence measurements. The obtained fluorescence and growth measurements will be analyzed and modeled accordingly. | The characterization process of the hydrocarbon sensing sub-project will entail output measurements of fluorescence at varying octane concentrations (0.1% – 1%) of the abovementioned strains. These measurements will be performed using a 96-well plate reader. Growth curves will be followed by OD-600 alongside the fluorescence measurements. The obtained fluorescence and growth measurements will be analyzed and modeled accordingly. | ||
| - | |||
| - | |||
Revision as of 12:03, 12 August 2010
Contents |
Genetic Regulation through Hydrocarbon Sensing
Aim
Creation of a BioBrick incorporating a regulatory mechanism controlled by alkane levels. High alkane levels will then be used to activate expression of specific genes involved in the alkane degradation. The BioBricks will be implemented in Escherichia coli K12 and characterized and evaluated on it regulating capabilities controlled by alkane levels.
Proposed Method
Based on: Canosa, I., J. M. Sanchez-Romero, et al. A positive feedback mechanism controls expression of AlkS, the transcriptional regulator of the Pseudomonas oleovorans alkane degradation pathway. Molecular Microbiology 35(4): 791-799 (2000)
Moreno, R., A. Ruiz-Manzano, et al. The Pseudomonas putida Crc global regulator is an RNA binding protein that inhibits translation of the AlkS transcriptional regulator. Molecular Microbiology 64(3): 665-675 (2007)
van Beilen, J. B., S. Panke, et al. Analysis of Pseudomonas putida alkane-degradation gene clusters and flanking insertion sequences: evolution and regulation of the alk genes. Microbiology-Sgm 147: 1621-1630 (2001)
Weissenborn, D. L., N. Wittekindt, et al.. "Structure and Regulation of the Glpfk Operon Encoding Glycerol Diffusion Facilitator and Glycerol Kinase of Escherichia coli K-12." Journal of Biological Chemistry 267(9): 6122-6131 (1992)
Functionality
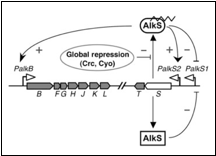
To control when the organism produces enzymes needed for the degradation of alkanes we will characterize an alkane sensing mechanism. The sensing mechanism proposed is that of the Pseudomonas putida GPo1 OCT plasmid alk genes cluster (figure 2). In the presence of alkanes, the AlkS transcriptional regulator activates the expression of its own gene, and that of alkT, from a promoter named PalkS2. This allows achieving AlkS levels that are high enough to activate the expression of the alkBFGHJKL operon from the PalkB promoter. AlkS recognizes C5–C10 n-alkanes as effectors, but does not respond to shorter or larger alkanes (Rojo et al. 2009)
The Crp-protein is a global regulator ubiquitous to E.coli which is known to bind to regions of promoters known to activate genes involved in the degradation of non-glucose carbon sources, in this way activating the genes downstream of it. We would like to seamlessly integrate the expression of our alkane degrading genes into the E.coli system by making the alkane-degrading genes sensitive to Crp.
Modelling
The modelling of the sensing system is described here
STEP 1: PalkS / AlkS BioBrick
Aim Create a construct suitable for the analysis of transcriptional activity over the PalkS promoter unit and insertion thereof into competent E.coli K12.
The first construct would be to analyze the activity of the PalkS1-2 promoter unit in the absence and presence of the active transcription factor, AlkS. AlkS is known to activate the alkane degradation genes operon (Alk genes) on the OCT plasmid in the presence of hydrocarbons. When AlkS binds to a hydrocarbon, it is activated and able to bind to the PalkS1 promoter region, inducing PalkS2 and increasing the transcription activity of its own gene, AlkS. In the absence of hydrocarbons, the AlkS transcription factor is inactive and its binding to PalkS1 represses transcription of the AlkS gene, thus keeping the AlkS protein at basal levels. In order to quantify these descriptions, a GFP translational unit will be placed behind the AlkS gene, thus making it dependent of the PalkS promoter unit. Fluorescence analyses in the absence and presence of hydrocarbons will provide us with the variations of AlkS transcription levels in the system.
Step 2: PalkS / AlkS / PalkB BioBrick
Aim To construct a plasmid containing the AlkS-PalkS-PalkB regulatory mechanism coupled to GFP and RFP generators in order to determine transcriptional activities of PalkS and PalkB at varying hydrocarbon concentrations by measuring fluorescence.
After having determined the transcriptional activity profile of the PalkS promoter unit, an analysis will be made of the transcriptional activity of the PalkB promoter. For the sake of transformational simplicity the PalkB region will be placed downstream of the AlkS CDS, thus on the same plasmid containing Construct A. The transcription initiation activity of PalkB will be analyzed by placing a RFP generator downstream of it. By having both the GFP and RFP profiles proper analyses can be performed concerning AlkS levels and PalkB activity at varying hydrocarbon concentrations.
STEP 3: P(CaiF) / ALKS / PalkB BioBrick
Aim The formation of a construct which facilitates the analysis of the transcriptional activity of the circuit responsive to hydrocarbons and to Crp.
Having analyzed the transcription activities of the PalkB / AlkS promoter units the following step is the analysis of the Crp-protein regulation mechanism. Analysis of the GFP levels which are coupled to AlkS expression will give the information needed on functionality of the circuit.
STEP 4: Characterization
Strains:
- PalkS / AlkS: E.coli K12/310C
- PalkS / AlkS / PalkB: E.coli K12/311C
- P(CaiF) / AlkS / PalkB: E.coli K12/312C
- Negative control: E.coli K12
The characterization process of the hydrocarbon sensing sub-project will entail output measurements of fluorescence at varying octane concentrations (0.1% – 1%) of the abovementioned strains. These measurements will be performed using a 96-well plate reader. Growth curves will be followed by OD-600 alongside the fluorescence measurements. The obtained fluorescence and growth measurements will be analyzed and modeled accordingly.
 "
"