Team:Stanford/Notebook/Lab Work/Week 4
From 2010.igem.org
Karpadillo (Talk | contribs) (→Karina's Notebook) |
Karpadillo (Talk | contribs) (→Karina's Notebook) |
||
| Line 487: | Line 487: | ||
5 uL Reverse primer<br/> | 5 uL Reverse primer<br/> | ||
40 uL PCR Supermix<br/><br/> | 40 uL PCR Supermix<br/><br/> | ||
| - | Make 3 PCR reactions of each sRNA to test temperature gradient. | + | Make 3 PCR reactions of each sRNA to test temperature gradient. <br/> |
| + | {| | ||
| + | |sRNA 1 labeled as 1, | ||
| + | |sRNA 2 labeled as 2 | ||
| + | |- | ||
| + | |sRNA 1C labeled as 1C, | ||
| + | |sRNA 2C labeled as 2C | ||
| + | |- | ||
| + | |} | ||
| + | <br/> | ||
| + | {| border="1" | ||
| + | |55.4 ºC | ||
| + | |61.9ºC | ||
| + | |52.0ºC | ||
| + | |- | ||
| + | |1a | ||
| + | |1b | ||
| + | |1c | ||
| + | |- | ||
| + | |2a | ||
| + | |2b | ||
| + | |2c | ||
| + | |- | ||
| + | |1Ca | ||
| + | |1Cb | ||
| + | |1Cc | ||
| + | |- | ||
| + | |2Ca | ||
| + | |2Cb | ||
| + | |2Cc | ||
| + | |} | ||
===Francisco's Notebook=== | ===Francisco's Notebook=== | ||
Revision as of 19:51, 11 August 2010

| Home | Project | Applications | Modeling | Parts | Team | Notebook |
Spring: Brainstorming | Spring Meetings
Summer: Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Week 7 | Week 8 | Week 9 | Week 10 | Summaries
|
7/19 Monday
Alex's Notebook
Run diagnostic. Restreak/inoculate. Gel extracted B0014. PCR parts. Finish other primers.
Laura's Notebook
set up the following ligations from the gel extractions done by me, Karina, and Francisco on Friday, 7/14/10)
- Francisco ran and imaged diagnostic gel on Friday
| Ligation Recipe | |
|---|---|
| dH2O | none |
| vector (with terminators attached) | 5.0 uL |
| insert (RFP or GFP) | 12.0 uL |
| 10X buffer | 2.0 uL |
| T4 ligase | 1.0 uL |
- normally run for 10 minutes at room temperature, overnight this time (started at 11:30 am)
Nanodrop data (deemed unreliable based on 260/230, possibly due do residual EtBr contamination)
| part | 260/280 | 260/230 | ng/uL |
| vector/terminator | 1.77 | 0.03 | 19.1 |
| RFP | 1.86 | 0.02 | 16.2 |
| GFP | 1.83 | 0.02 | 14.8 |
Karina's Notebook
Goal: Laura will ligate GFP and RFP and ligate them to terminators. We received our RSID + RBS oligos in the mail, so I will work on the PCR. I first need to make freezer stock and working stock of oligos. Won't start PCR until after lunch because we'll leave them overnight with Chris' PCR reactions.
Make Tris-HCl
Need .01 L of 10mM Tris-HCl solution. So, add 15.76 mg Tris-HCl to 10 mL H20.
- Hard to weigh out 15.8 mg, so instead got to 18.3 mg.
- Determined that need to add this to 11.6 mL H20
Make Freezer Stock of Oligos
Want 100 uM solution of Tris-HCl solution
- Amount of Tris-HCl to add depends on how much of the oligo's we received.
| amount (nmol) | mass (mg) | amount of Tris to add (uL) | |
| RSID 1 + RBS Forward | 70.2 | 1.82 | 702 |
| RSID 1 + RBS Reverse | 76.7 | 1.85 | 767 |
| RSID 2 + RBS Forward | 81.1 | 2.22 | 811 |
| RSID 2 + RBS Reverse | 83.5 | 2.09 | 835 |
Make Working Stock
Want a 10uM working stock solution
- 900 uL water + 100 uL Freezer Stock Solution
PCR Assembly
PCR Protocol calls for:
1.25 uL reverse primer
1.25 uL forward primer
DNA Template
50 uL PCR supermix
- For PCR Assembly, do not need DNA template. Also, add 4 times as much of each primer for PCR assembly.
Revised Recipe
5 uL Forward Primer
5 uL Reverse Primer
40 uL PCR Supermix
- we'll be using awesome PCR supermix- KEEP ON ICE
- Ran PCR with Chris, left running overnight
Francisco's Notebook
- Helped Karina and Laura
7/20 Tuesday
Alex's Notebook
1. Pick an isolated colony from an LB plate and grow overnight in 3–5 ml of LB at 37C. 2. Next morning, add 0.5 ml of the culture to 25 ml of LB in a 250-ml flask and grow at 37C to an OD600 of 0.50–0.60. 3. Transfer the culture to a 50-ml Falcon tube and spin at 6,000g in prechilled rotor for 10 min at 4C. 4. Wash the cell pellet with 20 ml of ice-cold H2O then centrifuge again as above. 5. Resuspend the pellet in 1 ml of H2O and transfer to a chilled 1.5-ml tube. Spin at 10,000g for 30 seconds at 4C. 6. Wash the cells again with 1 ml of ice cold H2O and centrifuge as above. 7. Repeat the above wash and spin step. 1. Resuspend the cell pellet in H2O in a final volume of 100μl and keep on ice.
Made LB. Lots of it. Check primers for MT. Make more competent cells. Digested PCR products (E/P). Ran most of the gel. Gel extract. I0500+B0034, F2620+B0034, J23107+B0034, AfsS C, and AfsS N. Ligate onto 1A2. Transform, with the old samples as well. Ran another PCR: 3,3,5,5,5.
Laura's Notebook
helped Alex with preparation of competent cells
- see protocol: [http://openwetware.org/wiki/Stanford/BIOE44:Module_1:Day3 Preparing Electrocompetent Cells]
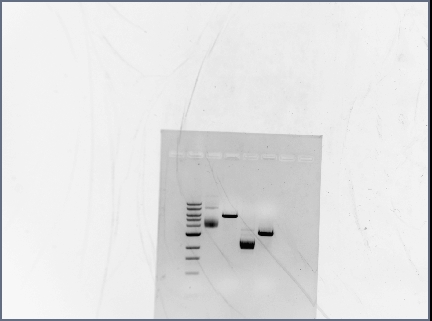
ran diagnostic gel for PCR-assembled DNA (Karina set up PCR rxns yesterday)
- order on gel:
- 100 bp ladder
- RSID1/RBS
- RSID2/RBS
Karina's Notebook
Goals: Run gel of PCR Assembly Product, gel extraction, digest, then run PCR clean up.
Make Gel
- Chris made me an 0.8% agarose gel
Load Gel
- added 10 uL loading dye directly into PCR tubes
- loaded 40 uL of each sample into wells
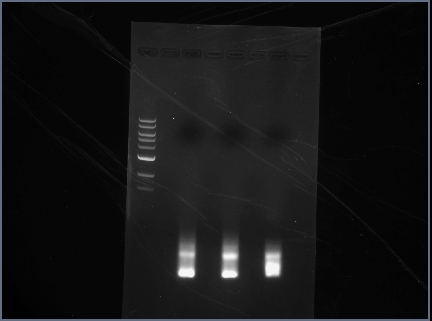
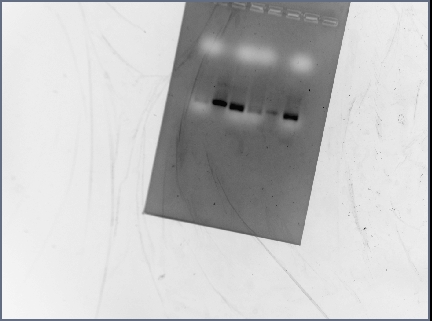
Gel Results
Analysis/Observations
Gel looked a little streaky to me, but we really only need the brightest bands (as circled). For next time:
- use 100 bp ladder (instead of 1 kb)
- Don't even bother with gel extraction after running a PCR. It is highly inefficient and a lot of DNA will be lost. Instead, run a a diagnostic gel to check that PCR assembly worked but then proceed to run a PCR cleanup and then digest.
- Gel with 100bp ladder:
Extraction
Extracted 1A, 1B, 2B, and 2C from the gel.
| sample | tube mass (g) | mass w/getl (g) | gel mass (g) | volume of QG to add (uL) |
| 1A | 1.01 | 1.25 | .24 | 720 |
| 1B | 1.01 | 1.28 | .27 | 810 |
| 2B | 1.01 | 1.24 | .23 | 690 |
| 2C | 1.01 | 1.24 | .23 | 690 |
- extracted and incubated each of the samples separately. To combine, loaded 1A and 1B into one spin column and 2B and 2C into another. Loaded up to 800 uL, spun down, loaded, then spun again. Then continued to follow [http://molecool.wustl.edu/krolllab/Kroll_Lab_Protocols/Molecular%20Biology%20protocols/Cloning%20protocols%20folder/Gel%20extraction-Qiagen.pdf procedure].
PCR Assembly
Set up another PCR assembly of RSID 1 and RSID 2 so that we have samples that have not been gel extracted. Ran another three samples of each. Will let run overnight.
- Primers were designed to anneal at 55º, but set up a temperature gradient to test annealing at other temperatures.
| RSID | temperature (ºC) |
| 1A | 68 |
| 1B | 63 |
| 1C | 56 |
| 2A | 68 |
| 2B | 63 |
| 2C | 56 |
Greg's Notebook
- Miniprepped pBAD, F2620, pSB1a2, pSB3c5
- Ran nanodrop diagnostic:
| Part # | 280 | 230 | concentration (ng/uL) |
| pBAD | 1.95 | 1.33 | 54.7 |
| F2620 | 1.88 | 1.48 | 185.1 |
| pSB3c5 | 1.71 | 0.85 | 40.4 |
| pSB1a2 | 1.81 | 1.38 | 153.4 |
- Poured large gel
- Helped Chris with electroporation transformation
7/21 Wednesday
Alex's Notebook
Redo ligations (3,7,7,2,1). Transform. Plate. Also run control. PCR: clean up, digest (FUCK UP: CUT w/ EP???), and gel electrophoresis (check lengths of DNA), ligate, and transform. Tomorrow. Diagnostic: I0500 (1255 bp) Good F2620 (1106 bp) Bad => primers were incorrect. J107 (98 bp) Not seen. Not seen again either on second try (1, 1.5, 2 ul template, working, stock). AfsS C (123 bp) Good AfsS NC (104 bp) Good
Redid PCR for J107 (6): 1,1.5,2 template, w/ stock or working primers. Made more RDs: 1A2 (2079 – 1069, EP), 3C5 (2738 – 1069, EP), T9 (1945, use BglI, 936 and 1221, EP), and (also use natural minipreped stuff, just diluted, on 1A3, to check transformation growth) F2620 (1061 – 2079, ES) (also cut at just SP, since on 1A2) E24 (876 – 2079, ES) and (also cut at just SP, since on 1A2). Mistake. Minipreped more cultures. Use above two for ligations to avoid red shit.
Laura's Notebook
today's digestions:
- RSID1, RSID2 (gel extracted from first PCR done by Karina)- digest with XbaI, PstI
- run overnight; started at 11:30am
- promoters within backbones (I0500-from Greg's box, and F2620-from Chris' box)- digest with SpeI, then PstI (done by Francisco)
- SpeI digest run for 3 hours, then heat killed 20 min. at 80oC, then PstI added overnight (to reduce enzyme competition for sites on the DNA, since recognition sequences are very close to each other)
Recipe:
| component | amount (uL) |
| DNA | 12.0 |
| H2O | 26.0 |
| 10X NEB buffer #2 | 5.0 |
| 10 X BSA | 5.0 |
| each enzyme | 1.0 (2.0 total) |
Karina's Notebook
Goal: Left Laura to gel extract previous PCR product as I worked on the new PCR product. Today, must make and run a diagnostic gel, PCR clean up, then digest RSID 1 and RSID 2.
Make TAE
Need a 1 L solution; we have 50x TAE stock
- 20 mL 50x TAE + 980 mL H20
Make Gel
Want a 2% agarose gel because oligos are short (~220 bp).
- 1 g agarose + 50 mL TAE
- 10 uL EtBr
Also, made Chris an 0.8% agarose gel (to repay him for yesterday).
Load Gel
10 uL 100 bp ladder
1 uL dye + 1 uL DNA
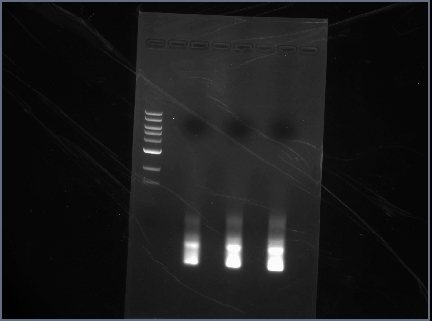
Results
Great! No difference in the annealing temperatures. All bands are clear and bright, can use all three samples for restriction digests.
PCR Cleanup
Used Qiagen MinElute PCR Purification Kit Protocol.
- 50 uL reaction, therefore use 250 uL buffer PB
- Followed protocol and eluted in 10 uL water.
Restriction Digest
Cut at X and P
Recipe
- 10 uL DNA
- 5 uL NEBuffer 2
- 5 uL BSA
- 1 uL Xba
- 1 uL Pst
- 28 uL H20
Total volume: 50 uL
Let run overnight in 37º waterbath
Francisco's Notebook
- Digested RSID+RBS's and promoter backbones
- RSID1+RBS: digested with Xba1 and Pst1
- RSID2+RBS: digested with Xba1 and Pst1
- pBad (I0500): digested with Spe1, then heat inactivated, then digested with Pst1^
- pLux (F2620): digested with Spe1, then heat inactivated, then digested with Pst1^
^The restriction enzymes were added one after the other so that they would not compete when binding to nearby stretches of DNA.
- Made 100uL of 10X BSA stock from 100X BSA stock.
- Laura ligated GFP and RFP inserts to terminator backbones on Monday. Ligated terminator backbones to itself (no insert added) as a control to check that the terminator backbone was cut at both E and X.
Greg's Notebook
- Ran restriction digest of miniprepped DNA
- Inoculated 3 tubes each with 5 mL LB and F2620, T9002, pSB3c5, pSB1a2, I0500, E0240
- Gel-extracted some stuff for Alex
- Did two electroporations
7/22 Thursday
Alex's Notebook
Do above. Check plates. Re-streak some. Gel electrophoresis for miniprep restriction digest. Gel extraction and ligation. Gel electrophoresis for restriction digest of PCR stuff. Gel extraction and ligation. PCR cleanup for J107, check with Isis/Ryan first. Inoculated J23100, J23107, and transformed J23119. P1, 18A, Plasmid: pSB1A2
Make: .8% gel with 15 wells. Run green. 3% gel with 15 wells. Run blue.
Laura's Notebook
- did gel extraction (Qiaquick Gel Extraction Kit) on the following samples:
| part/digestion | mass tube (g) | mass tube + gel (g) | mass gel (g) | uL QG to add |
| I0500 (pBAD) | 1.00 | 1.06 | 0.06 | 180 |
| F2620 | 1.00 | 1.05 | 0.05 | 150 |
| RSID1KP | 1.00 | 1.05 | 0.05 | 150 |
| RSID1LEO | 1.01 | 1.08 | 0.07 | 210 |
| RSID2KP | 1.00 | 1.07 | 0.07 | 210 |
| RSID2LEO | 1.01 | 1.05 | 0.04 | 120 |
- RSID1KP and RSID2KP- PCR done by Karina, then gel extracted prior to digestion, digestion done by Karina
- RSID1LEO and RSID2LEO- PCR done by Karina, digested by Laura
- ran on 1.5% gel at 75V for 1 hour
- 10uL ladder (1KB)
- 1uL each sample + 1uL loading dye- in order on chart
Gels used for gel extraction
- Digested RSID's:
- Digested Promoters:
Made 4 0.5L bottles of agar
- recipe:
- 5 g bacto-peptone
- 2.5 g yeast extract
- 2.5 g NaCl
- 6 g agar
- 485 mL dH2O (did first bottle in graduated cylinder, powders shifted volume by 15 mL- 450 mL went up to 465 mL, mixed and added to bottle, then used 35 mL dH20 to rinse cylinder contents into bottle)
Karina's Notebook
Goal: Laura did gel extraction of RSIDs, F2620, and I500 this morning. We'll need to ligate the IDs to the promoters then transform, but must wait until Francisco gel etracts promoters. In the mean time, I helped Chris with his PCR cleanup:
| Samples | Volume | PB To add (uL) |
| G1 | 50 | 250 |
| G2 | 50 | 250 |
| G3 | 50 | 250 |
| LM (X/P) | 50 | 250 |
For G1, G2, and G3, elute in 50 uL H20.
For LM X/P elute in 30 uL H20. Because LM is ready for ligation, eluting in 30 uL makes it more concentrated.
RECIEVED OLIGOS!
make 100 uMol stock
| Primer (F or R) | nmol | TRIS-HCL to add (uL) | |
| sRNA_1 | F | 4 | 40 |
| sRNA_1 | R | 4 | 40 |
| sRNA_2 | F | 4 | 40 |
| sRNA_2 | R | 2.7 | 33 |
| sRNA_1C | F | 4 | 40 |
| sRNA_1C | R | 3.3 | 33 |
| sRNA_2C | F | 4 | 40 |
| sRNA_2C | R | 4 | 40 |
Make 10 uMol Working Stock
90 uL H20 + 10 uL Oligo Stock
Set up PCR of sRNAs
Recipe:
5 uL forward primer
5 uL Reverse primer
40 uL PCR Supermix
Make 3 PCR reactions of each sRNA to test temperature gradient.
| sRNA 1 labeled as 1, | sRNA 2 labeled as 2 |
| sRNA 1C labeled as 1C, | sRNA 2C labeled as 2C |
| 55.4 ºC | 61.9ºC | 52.0ºC |
| 1a | 1b | 1c |
| 2a | 2b | 2c |
| 1Ca | 1Cb | 1Cc |
| 2Ca | 2Cb | 2Cc |
Francisco's Notebook
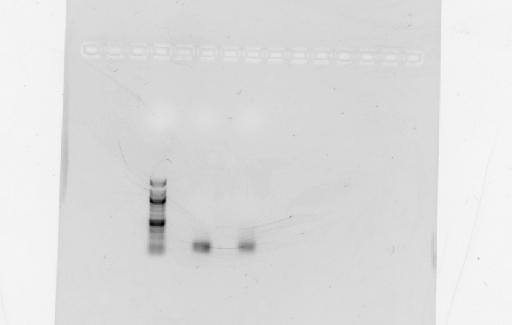
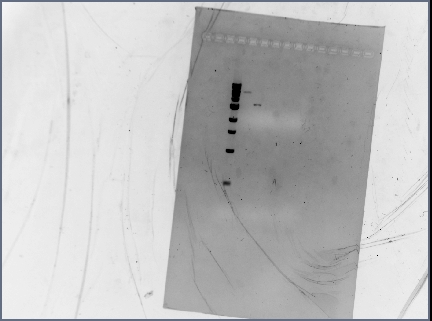
- Digested RSID's did not show up in the diagnostic gel:
- Rerun diagnostic gel for the RSID's, using high percentage gel and 100kb ladder:
- Possible problems:
- Gel extraction has low yield. Plan for next time: re-run PCR of RSID's, but forgo gel extract if PCR reaction is clean.
- Not enough DNA was loaded in the gel. Plan for next time: try loading more volume of DNA
- We have prepared GFP-terminator and RFP-terminator ligations on Monday, and a control ligation with the terminator backbone and no insert on Wednesday. Hope to transform bacteria with those ligations soon.
- Re-aliquoted 1000uL of electrompetent cells Alex prepared into 20 tubes of 50uL each. Stored in 80 deg C freezer and will start transformations on Friday.
7/23 Friday
Friday Recap Meeting
- Leader: Francisco
Alex's Notebook
Ran e0240 and i0500 on .8% gel. PCR RD => do MinElute and record concentration, ligate, and transform as appropriate (DH5a). Run diagnostic; determine which is which and check for brightness. Primers. Redo PCR; gradient. Sequence J23107.
Laura's Notebook
designed primers for colony PCR
tips for primer design (from Smolke Lab, via Chris VanLang)
- 18-24 bases
- ~60oC annealing temperature
- avoid: high or low GC content, repeats of >3 bases
- for use in later sequencing reactions, place primers ~ 50 bp outside region of interest
primer sequences:
pBADfor: gattagcggatcctacctgacgc
2K3rev: GGAAGCCTGCATAACGCGAAGT
F2620for: gggtgggcctttctgcgtttatat
IA2rev: AGTGAGCTGATACCGCTCGC
pairs:
- pSB2K3, I0500 (pBAD)
- pSB1A2, F2620 (pLUX)
Francisco's Notebook
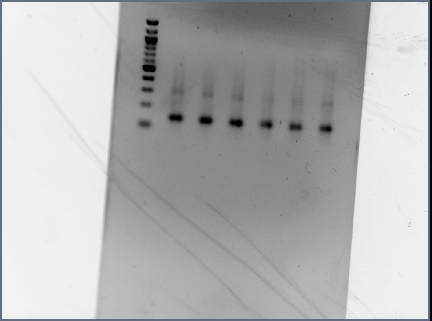
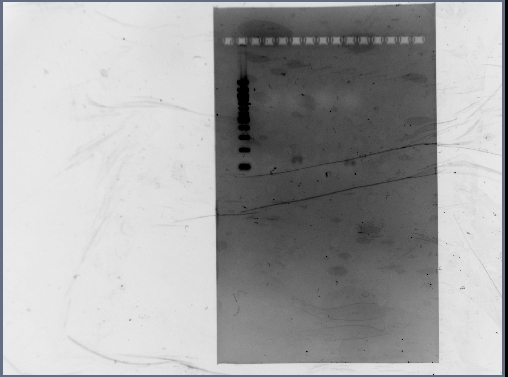
- Reran PCR assembly (30 cycles of 'YA' program) of RSID's+RBS
- ran three reactions for RSID1+RBS (tubes labelled 1a, 1b, 1c) and RSID2+RBS (tubes labelled 2a, 2b, 2c)
- annealing temperatures (deg C): 57.0, 55.2, 53.7 for a, b, c respectively.
- gel results looked good:
- Transformed E. coli BW strain with ligations prepared earlier in the week: GFP-terminator, RFP-terminator, and terminator ligated with no insert (negative control). 0.5 uL of DNA was used for each transformation and the settings on the zapper was 2000 V, 25 uF, 200 ohms. Some of the cells sparked and had to be discarded. Incubated for almost an hour after electroporation, spun down the cells at 3000g for 3 min. Transferred cells to plates, and left the plates to incubate overnight.
- Saturday morning update: None of the plates had colonies. =(
7/24 Saturday
Alex's Notebook
Inoculated 100, 107, and 119 at 2:10 pm
Ligations: A few. 4, 6.5, 6.5, 2, 1 or 4, 6, 2, 1, 7 H2O. 3C5: T9 and I5+E24 In progress 1A2: T9 and I5+E24 In progress
PCR: sGFP In progress ATEAM: 45 supermix, 1.5 ul primers, and 2 ul template (I746908 2093 bp).
DpnI: get rid of template Cuts: GAmeTC/ CTAmeG Reacts: 37 C for 1 hour Denatures: 20 min at 80 C Nanodrop Diagnostic gel (883 bp)
PCR purify Nanodrop
Restriction Digest Some at EP and some at XP Nanodrop
Enz clean up Nanodrop
Promoters Freezer stock: 100 uM (use 10 mM Tris-HCL) Working stock: 20 uM (5x dilution) Std. PCR: 45 supermix, 500 nM primers (1.25 ul from 20 uM), 250 ng (varies by concentration) template, variable H2O. Stiching PCR: 40 supermix, 5 ul each promoter (2000 nM). Try: 45sm, 1.25 ul each (500 nM) and 3.75 H2O. Gel: 12 (40), 34, 12 (45), 34, 1.25, .625, O4, O5, C3, C6, N3, and N5
To be done: I5+RBS: 160 ul total, split into 8x 20 ul. 144 ul of supermix, 8 ul (2x 4 ul) primers, 800 ng template (),
variable H2O.
Grad (50-60): 50, 50.8, 52.1, 54, 56.2, 58, 59.3, 60 F26+RBS Primers not here.
J100+RBS Template not ready until 3 am.
J107+RBS Template unconfirmed. Wait until 3 am.
In progress:
Afs C: 160 ul total, split into 8x 20 ul. 144 ul of supermix, 8 ul (2x 4 ul) primers.
Grad (50-60), 5 cycles
Conc. Fwd: .027 umoles
Rev: .023 umoles
Afs NC: 160 ul total, split into 8x 20 ul. 144 ul of supermix, 8 ul (2x 4 ul) primers.
Grad (50-60), 5 cycles
Conc. Fwd: .030 umoles
Rev: .030 umoles
Afs 4: 160 ul total, split into 8x 20 ul. 144 ul of supermix, 8 ul (4x 2 ul) primers. Original AfsS promoter. Grad (50-60), 5 cycles Conc. 1: .027 umoles 2: .029 umoles 3: .029 umoles 4: .027 umoles
Inoculate: All on Amp 100 In progress 107 In progress 119 In progress
Digest: B0034 at XP In progress
J100 at ES Not begun J107 at ES Not begun J119 at ES Not begun
Ligate into 3C5 Screen C first, then Amp to confirm one plasmid present.
Gel for PCR purification Ones that got f-ed up. In progress.
 "
"