Team:TU Delft/27 July 2010 content
From 2010.igem.org
(→Alkane degradation) |
|||
| Line 9: | Line 9: | ||
|'''Expected length (bp)''' | |'''Expected length (bp)''' | ||
|'''Status''' | |'''Status''' | ||
| - | |||
|- | |- | ||
|1 | |1 | ||
| Line 15: | Line 14: | ||
|n/a | |n/a | ||
|n/a | |n/a | ||
| - | |||
|- | |- | ||
|2 | |2 | ||
| Line 21: | Line 19: | ||
|198, | |198, | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|- | |- | ||
|3 | |3 | ||
| Line 27: | Line 24: | ||
| | | | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|- | |- | ||
|4 | |4 | ||
| Line 33: | Line 29: | ||
|207, | |207, | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|- | |- | ||
|5 | |5 | ||
| Line 39: | Line 34: | ||
| | | | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|- | |- | ||
|6 | |6 | ||
| Line 45: | Line 39: | ||
|1230 | |1230 | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|- | |- | ||
|7 | |7 | ||
|undigested rubR | |undigested rubR | ||
| - | |||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| | | | ||
| Line 57: | Line 49: | ||
|1350, | |1350, | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|- | |- | ||
|9 | |9 | ||
| Line 63: | Line 54: | ||
| | | | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|- | |- | ||
|10 | |10 | ||
| Line 69: | Line 59: | ||
|777, | |777, | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|- | |- | ||
|11 | |11 | ||
| Line 75: | Line 64: | ||
| | | | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|- | |- | ||
|12 | |12 | ||
| Line 81: | Line 69: | ||
|1521 | |1521 | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|- | |- | ||
|13 | |13 | ||
| Line 87: | Line 74: | ||
| | | | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|- | |- | ||
|14 | |14 | ||
| Line 93: | Line 79: | ||
|2116 | |2116 | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|- | |- | ||
|15 | |15 | ||
| Line 99: | Line 84: | ||
| | | | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|- | |- | ||
|16 | |16 | ||
| Line 105: | Line 89: | ||
|2116 | |2116 | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|- | |- | ||
|17 | |17 | ||
| Line 111: | Line 94: | ||
| | | | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|- | |- | ||
|18 | |18 | ||
| Line 117: | Line 99: | ||
|2116 | |2116 | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|- | |- | ||
|19 | |19 | ||
| Line 123: | Line 104: | ||
| | | | ||
|<font color=limegreen>✓</font> | |<font color=limegreen>✓</font> | ||
| - | |||
|} | |} | ||
Revision as of 20:32, 5 August 2010
Alkane degradation
Yesterday's digestion products were checked on a gel.
Lane description:
| # | Description | Expected length (bp) | Status |
| 1 | SmartLadder | n/a | n/a |
| 2 | rubA3 + XbaI + PstI + PvuI | 198, | ✓ |
| 3 | undigested rubA3 | ✓ | |
| 4 | rubA4 + XbaI + PstI + PvuI | 207, | ✓ |
| 5 | undigested rubA4 | ✓ | |
| 6 | rubR + XbaI + PstI + EcoRI | 1230 | ✓ |
| 7 | undigested rubR | ✓ | |
| 8 | ladA + XbaI + PstI + PvuI | 1350, | ✓ |
| 9 | undigested ladA | ✓ | |
| 10 | ADH + XbaI + PstI + EcoRI | 777, | ✓ |
| 11 | undigested ADH | ✓ | |
| 12 | ALDH + XbaI + PstI + PvuI | 1521 | ✓ |
| 13 | undigested ALDH | ✓ | |
| 14 | J61100 + SpeI + PstI | 2116 | ✓ |
| 15 | undigested J61100 | ✓ | |
| 16 | J61101 + SpeI + PstI | 2116 | ✓ |
| 17 | undigested J61101 | ✓ | |
| 18 | J61107 + SpeI + PstI | 2116 | ✓ |
| 19 | undigested J61107 | ✓ |
As can be seen from the gel, all digestions went well, let's hope that the ligation / transformation also go well!
Transformation
The ligation mixes were incubated overnight. We transformed 10 μL of these ligations in Top10 competent cells, and these were grown on LB-plates with antibiotic over night.
RBS Characterization
For our RBS characterization project, 5 different RBS sequences from the [http://partsregistry.org/Ribosome_Binding_Sites/Prokaryotic/Constitutive/Anderson Anderson RBS family] where placed in front of GFP and measured over 18 hours using a Gen5 fluorenscence and absorbance plate reader
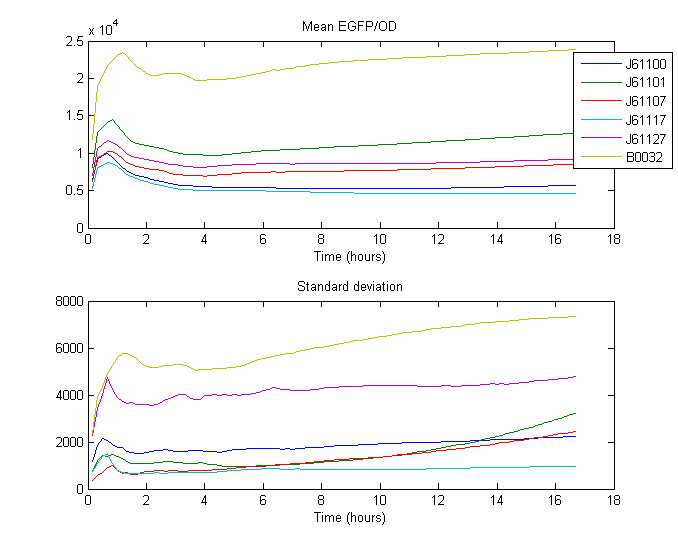
Strength was calculated by taking the mean of the ratio between the expression of known RBS ([http://partsregistry.org/Part:BBa_B0032 B0032]) and expression of Anderson RBS over some time. Expression being the measured fluorescence divided by measured biomass (absorbance, OD).
The measurements where taken from the part of the curve where optimal growth can be assumed, so from 0:40 until 2:30
| RBS | Strength |
| [http://partsregistry.org/Part:BBa_J61100 J61100] | 0.047513 |
| [http://partsregistry.org/Part:BBa_J61101 J61101] | 0.119831 |
| [http://partsregistry.org/Part:BBa_J61107 J61107] | 0.065454 |
| [http://partsregistry.org/Part:BBa_J61117 J61117] | 0.038518 |
| [http://partsregistry.org/Part:BBa_J61127 J61127] | 0.087334 |
| [http://partsregistry.org/Part:BBa_B0032 B0032] | 0.300000 |
Next step: We want to relate the RBS sequence to the strength, so it might be possible to say predict something about the other Anderson RBS sequences.
 "
"
