Team:TU Delft/8 July 2010 content
From 2010.igem.org
(Difference between revisions)
| Line 22: | Line 22: | ||
|1158 | |1158 | ||
|G00100 + G00101 | |G00100 + G00101 | ||
| - | |<font color=limegreen> | + | |<font color=limegreen>✓</font> |
|Unexpected band at 2700 | |Unexpected band at 2700 | ||
|- | |- | ||
| Line 29: | Line 29: | ||
|1158 | |1158 | ||
|G00100 + G00101 | |G00100 + G00101 | ||
| - | |<font color=limegreen> | + | |<font color=limegreen>✓</font> |
| | | | ||
|- | |- | ||
| Line 36: | Line 36: | ||
|1158 | |1158 | ||
|G00100 + G00101 | |G00100 + G00101 | ||
| - | |<font color=limegreen> | + | |<font color=limegreen>✓</font> |
| | | | ||
|- | |- | ||
| Line 43: | Line 43: | ||
|1158 | |1158 | ||
|G00100 + G00101 | |G00100 + G00101 | ||
| - | |<font color=limegreen> | + | |<font color=limegreen>✓</font> |
| | | | ||
|- | |- | ||
| Line 50: | Line 50: | ||
|1158 | |1158 | ||
|G00100 + G00101 | |G00100 + G00101 | ||
| - | |<font color=limegreen> | + | |<font color=limegreen>✓</font> |
| | | | ||
|- | |- | ||
| Line 57: | Line 57: | ||
|1158 | |1158 | ||
|G00100 + G00101 | |G00100 + G00101 | ||
| - | |<font color=limegreen> | + | |<font color=limegreen>✓</font> |
| | | | ||
|- | |- | ||
| Line 64: | Line 64: | ||
|1158 | |1158 | ||
|G00100 + G00101 | |G00100 + G00101 | ||
| - | |<font color=limegreen> | + | |<font color=limegreen>✓</font> |
| | | | ||
|- | |- | ||
| Line 71: | Line 71: | ||
|1158 | |1158 | ||
|G00100 + G00101 | |G00100 + G00101 | ||
| - | |<font color=limegreen> | + | |<font color=limegreen>✓</font> |
|probably run of the gel | |probably run of the gel | ||
|- | |- | ||
| Line 78: | Line 78: | ||
|1158 | |1158 | ||
|G00100 + G00101 | |G00100 + G00101 | ||
| - | |<font color=limegreen> | + | |<font color=limegreen>✓</font> |
| | | | ||
|- | |- | ||
| Line 85: | Line 85: | ||
|1158 | |1158 | ||
|G00100 + G00101 | |G00100 + G00101 | ||
| - | |<font color=limegreen> | + | |<font color=limegreen>✓</font> |
|Sample not fully loaded on gel | |Sample not fully loaded on gel | ||
|- | |- | ||
Revision as of 18:46, 2 August 2010
Contents |
Lab work
Ordered DNA stocks
We harvested the 200 mL bacterial cells of the 16 DNA parts. We used 3 mL of the bacterial cells to make -80 °C stocks. With the rest we centrifuged at 4,000 rmp for 15 minutes and stored the pellets in the -20 °C.
Characterization of Anderson RBS sequences
Tuesday's ligation products of BioBricks K398500-K398504 yielded successful transformants. Single colony PCRs were performed and loaded onto a gel. The bands on the 1% agarose gel indicated the presence of inserts with the proper lengths:
Lane descriptions:
| # | Description | Expected Length (bp) | Primers | Status | Remarks |
| 1 | transformant #1 of ligation mix K398500 | 1158 | G00100 + G00101 | ✓ | Unexpected band at 2700 |
| 2 | transformant #2 of ligation mix K398500 | 1158 | G00100 + G00101 | ✓ | |
| 3 | transformant #1 of ligation mix K398501 | 1158 | G00100 + G00101 | ✓ | |
| 4 | transformant #2 of ligation mix K398501 | 1158 | G00100 + G00101 | ✓ | |
| 5 | transformant #1 of ligation mix K398502 | 1158 | G00100 + G00101 | ✓ | |
| 6 | transformant #2 of ligation mix K398502 | 1158 | G00100 + G00101 | ✓ | |
| 7 | transformant #1 of ligation mix K398503 | 1158 | G00100 + G00101 | ✓ | |
| 8 | transformant #2 of ligation mix K398503 | 1158 | G00100 + G00101 | ✓ | probably run of the gel |
| 9 | transformant #1 of ligation mix K398504 | 1158 | G00100 + G00101 | ✓ | |
| 10 | transformant #2 of ligation mix K398504 | 1158 | G00100 + G00101 | ✓ | Sample not fully loaded on gel |
| 11 | transformant #1 of ligation control | G00100 + G00101 | probably run of the gel | ||
| 12 | transformant #2 of ligation control | G00100 + G00101 | |||
| 13 | transformant #1 of digestion control | G00100 + G00101 | probably run of the gel | ||
| 14 | transformant #2 of digestion control | G00100 + G00101 | |||
| M1 | SmartLadder Marker | n/a | n/a | n/a |
The colonies belonging to lanes 2, 4, 6, 8 and 10 were plated out on AMP plates to yield reincultures for subsequent characterization experiments.
Alkane degradation
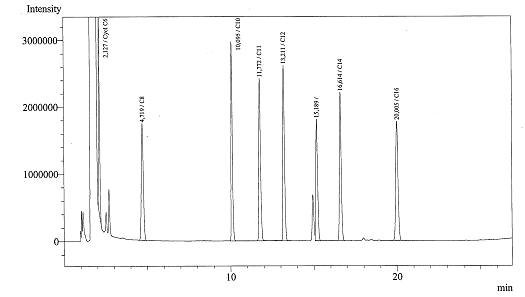
Today we got the Gas Chromatograph working, we could identify several peaks. Now we are ready for the real experiments!
 "
"