Team:Stockholm/Planning
From 2010.igem.org
m (→Protein A) |
m (→Melanin) |
||
| Line 71: | Line 71: | ||
===Melanin=== | ===Melanin=== | ||
| + | |||
| + | In the “normal“ skin melanocytes are present and secrete out melanin molecules which enter the keratocytes, resulting in color in the skin. In “vitiligo“ skin melanocytes are missing at some areas, thus no melanin production takes place at these patches that keratocytes can take up and result in skin color, instead one gets white patches. Note! Ignore albinism and piebaldism. | ||
| + | |||
| + | [[Image:Melanin1.png]] | ||
| + | |||
| + | Our aim is to have our bacteria on the skin to produce the missing melanin in vitiligo skin as a fusing protein with a cell penetrating peptide, thus passing into the skin layer (epidermis) and entering the “empty of melanin“ keratocytes. This will result in giving a vitiligo skin back its color somewhat directly (matter of days maybe, depending on how much melanin our bacteria produces and enters the target cells ->keratocytes). This approach of allowing for repigmentation to occur will help many people in a short time (compared to current techniques, taking 6 -12 months). Additionally, the vitiligo skin will be more tolerable against UV-light thanks to the bacteria produced melanin entering keratocytes. | ||
| + | |||
| + | We have found an article about transgenic e.coli producing one type of melanin. Hopefully in the future one might by research find a color spectrum of different pigment molecules (melanin) that will match to different people’s skin color. | ||
| + | |||
| + | ---- | ||
| + | |||
| + | ''“Melanins are polyphenolic heteropolymers with a wide range of colors and applications. Due to their chemical composition, melanins have physicochemical properties that allow them to act as ultraviolet absorbers, cation exchangers, drug carriers, amorphous semiconductors, X-ray and γ-ray absorbers, and in some instances, substrates with antioxidant and antiviral activity (Riley 1997; Lagunas-Muñoz et al. 2006). Bacteria are known to produce 3,4-dihydroxyphenylalanine-melanins, the pheomelanins or eumelanins, and melanin-like pigments that are derived from non-nitrogenous phenols (the allomelanins or pyomelanins; Gibson and George 1998).”'' - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melanin in Escherichia coli p. 125 | ||
| + | |||
| + | ---- | ||
| + | |||
| + | ''“Production of melanins by recombinant E. coli is useful in industrial processes.”'' - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melanin in Escherichia coli p. 125 | ||
| + | |||
| + | ---- | ||
| + | |||
| + | ''“The clone fss6 producing red-brown pigment was isolated and further characterized in the study. Based on the physico-chemical characteristics of the pigment, the red-brown pigment was identified as melanin.”'' - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melaninin Escherichia coli p. 129 | ||
| + | |||
| + | ---- | ||
| + | |||
| + | ''"[...] the production of melanin was correlated with homogentistic acid (HGA). The p-hydroxyphenylpyruvate produced by the Escherichia coli host was converted to HGA through the oxidation reaction of introduced HPPD."'' - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melaninin Escherichia coli p. 124 | ||
| + | |||
| + | ---- | ||
| + | |||
| + | ''"HPLC analysis for HGA from the cell-free filtrate of cultures showed that HGA was produced by recombinant E.coli strains before melanin was produced....HGA is the product of ORF2 and should be the precursor of melanin."'' - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melaninin Escherichia coli p. 129 | ||
| + | |||
| + | ---- | ||
| + | |||
| + | ''"The cloned gene (ORF2) encoding putative 4-hydroxyphenylpyruvate dioxygenase (HPPD) was responsible for the production of melanin in E. coli."'' - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melaninin Escherichia coli p. 129 | ||
| + | |||
| + | ---- | ||
| + | |||
| + | ''"The known pathway for the catabolism of tyrosine to HGA in mammals and bacteria is that tyrosine is converted to p-hydroxyphenylpyruvate (p-HPP) through a transamination reaction. Then, p-HPP is converted to HGA through the oxidation of HPPD (Lindstedt and Odelhög 1987; Lindstedt et al. 1977). In the study, the transaminase for the initial step should exist in clone fss6. Three aminotransferase capable of converting tyrosine into p-HPP have been identified in E. coli (Pittard 1987). We propose that tyrosine is first catabolized by aminotransferases of E. coli host, then, the produced p-HPP is converted to HGA by the introduced HPPD gene products. Lastly, the accumulation and polymerization of HGA results in the formation of melanin."'' - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melaninin Escherichia coli p. 130 | ||
===MITF=== | ===MITF=== | ||
Revision as of 15:02, 18 May 2010

| Home | Team | Project | Planning | Notebook | Protocols | Pictures | Journal club | Sponsors |
|---|
Planning
Here follows the ongoing discussion amongst the group members about the project we are putting together. In addition we include theoretical support from scientific articles for what we want to accomplish.
Cell penetrating peptide (CPP)
Protein A
Protein A would be secreted from the bacteria, penetrating the skin and bind IgG antibodies - that in vitiligo patients bind melanocytes - so that the igG antibodies can not induce an immune response.
"It binds proteins from many of mammalian species, most notably IgGs" - http://en.wikipedia.org/wiki/Protein_A (Find better source)
Johan: Good as IgGs are the main antibody in excess for vitiligo
“It binds with the Fc region of immunoglobulins through interaction with the heavy chain. The result of this type of interaction is that, in serum, the bacteria will bind IgG molecules in the wrong orientation (in relation to normal antibody function) on their surface which disrupts opsonization and phagocytosis.” - http://en.wikipedia.org/wiki/Protein_A
Johan: The disruption is what we want (are there more disruptions to think of, other than opsonization and phagocytosis?)
“It binds with high affinity to human IgG1 and IgG2 as well as mouse IgG2a and IgG2b. Protein A binds with moderate affinity to human IgM, IgA and IgE as well as to mouse IgG3 and IgG1.[1] It does not react with human IgG3 or IgD, nor will it react to mouse IgM, IgA or IgE.” - http://en.wikipedia.org/wiki/Protein_A
Johan: See if the antibody in excess in vitiligo hopefully is IgG1/IgG2
“... possesses several properties which makes it suitable as a partner in fusion proteins: (a) SpA enables purification of the fusion protein by affinity chromatography due to its specific binding to the Fc part of immunoglobulins, (b) it is competent for secretion in E. coli, (c) it is monomeric and relatively small, (d) it has been reported to be extremely soluble in water solutions (Samuels- son et al., 1991), (e) it is proteolytically very stable intracellularly (Nilsson et al., 1985a) and in the periplasm of E. coli (Nilsson and Abrahms6n, 1990). Fusions to the IgG-binding domains of SpA have successfully been applied for high-level production of peptide hormones (Abrahms6n et al., 1986; Moks et al., 1987a, b; Nilsson et al., 1991, 1985b) and as a tool for the production of specific antibodies against gene products (L6wenadler et al., 1987, 1986).” - Fusions to the 5' end of a gene encoding a two-domain analogue of staphylococcal protein A, pp 270-271
Johan: There are shortened versions of protein A. These might have some advantages. E.g. they dont have the domain that normally binds to the cell membrane, instead they are secreted extracellularly.
http://johan.nordholm.se/Skola/igem/Z.pdf Interesting information about the Z domain of Protein A, text from the doctoral thesis "Interaction-Engineered Three-Helix Bundle Domains for Protein Recovery and Detection" by Tove Alm from 2010 at KTH
Andreas: How will we prevent protein A from entering the blood stream? If it is able to penetrate the skin cells to the spot where antibodies are present, it is also likely to ”exit” the skin into the blood stream. If this happens, protein A will not only cause a severe immune shock, it will also interfere with any type of antibody in the blood, not just the ones targeting vitiligo. This is especially true if Protein A is proteolytically very stable, as stated in (e) above.
Are there any examples mammalian in vivo Protein A studies? It would be a great leap forward if there are any such examples, showing how Protein A interacts with the mammalian immune system. - I found a website at the US National Cancer Insitute, where they mention two clinical trials aiming at using Staphylococcus aureus Protein A as a cancer treatment. The clinical trials made it to phase 2 (out of the usual 4, if I remember correctly), but were then closed. I haven’t been able to find any statements, links or articles on why they were closed, but here is a link to the NCI website: http://ncit.nci.nih.gov/ncitbrowser/ConceptReport.jsp?dictionary=NCI%20Thesaurus&code=C17166 - Now I found an article from the mid-80s on Protein A. Not sure if it is connected to the clinical trials at NCI, but here’s a PubMed link: http://www.ncbi.nlm.nih.gov/pubmed/6379115 - There are also several articles mentioning ex vivo methods with Protein A for cancer treatments.
Nina: Size of protein A with Z domain: 7 kDA
IgG protease
IgG antibodies are enriched in vitiligo patients. IgG proteases could partially solve this problem. Also, IgG protease could be fused with Protein A to get more activity from the protease.
"Actinobacillus actinomycetemcomitans produces a protease to human immunoglobulin G that is an important evasion mechanism […] The results showed that A. actinomycetemcomitans produced a protease to human immunoglobulin G in the culture supernatant […] The molecular mass of the protease active fraction was from 43 to 150 kDa." - IMMUNOGLOBULIN G PROTEOLYTIC ACTIVITY OF ACTINOBACILLUS ACTINOMYCETEMCOMITANS
Johan: Their IgG protease eluted in fraction 13-17. +150 kDa eluted in fraction 9, and 43 kDa eluted in fraction 19. So at least it seems that the IgG protease is closer to 43 kDa than to +150 kDa.. (I wouldnt want to be a CPP carrying a cargo +150 kDa...)
"IdeS, a proteinase from Streptococcus pyogenes, cleaves immunoglobulin (Ig)G antibodies with a unique degree of specificity. Pathogenic IgG antibodies constitute an important clinical problem contributing to the pathogenesis of a number of autoimmune conditions and acute transplant rejection. To be able to effectively remove such antibodies is therefore an important clinical challenge." - IdeS: A Bacterial Proteolytic Enzyme with Therapeutic Potential
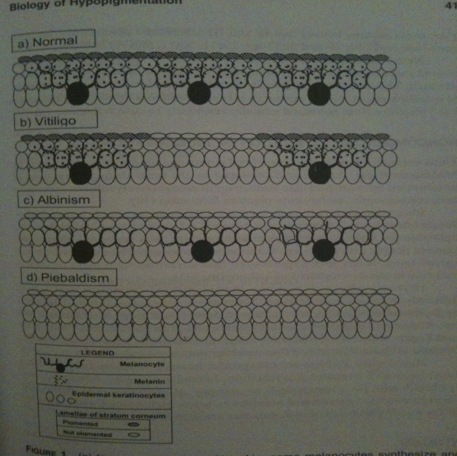
Melanin
In the “normal“ skin melanocytes are present and secrete out melanin molecules which enter the keratocytes, resulting in color in the skin. In “vitiligo“ skin melanocytes are missing at some areas, thus no melanin production takes place at these patches that keratocytes can take up and result in skin color, instead one gets white patches. Note! Ignore albinism and piebaldism.
Our aim is to have our bacteria on the skin to produce the missing melanin in vitiligo skin as a fusing protein with a cell penetrating peptide, thus passing into the skin layer (epidermis) and entering the “empty of melanin“ keratocytes. This will result in giving a vitiligo skin back its color somewhat directly (matter of days maybe, depending on how much melanin our bacteria produces and enters the target cells ->keratocytes). This approach of allowing for repigmentation to occur will help many people in a short time (compared to current techniques, taking 6 -12 months). Additionally, the vitiligo skin will be more tolerable against UV-light thanks to the bacteria produced melanin entering keratocytes.
We have found an article about transgenic e.coli producing one type of melanin. Hopefully in the future one might by research find a color spectrum of different pigment molecules (melanin) that will match to different people’s skin color.
“Melanins are polyphenolic heteropolymers with a wide range of colors and applications. Due to their chemical composition, melanins have physicochemical properties that allow them to act as ultraviolet absorbers, cation exchangers, drug carriers, amorphous semiconductors, X-ray and γ-ray absorbers, and in some instances, substrates with antioxidant and antiviral activity (Riley 1997; Lagunas-Muñoz et al. 2006). Bacteria are known to produce 3,4-dihydroxyphenylalanine-melanins, the pheomelanins or eumelanins, and melanin-like pigments that are derived from non-nitrogenous phenols (the allomelanins or pyomelanins; Gibson and George 1998).” - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melanin in Escherichia coli p. 125
“Production of melanins by recombinant E. coli is useful in industrial processes.” - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melanin in Escherichia coli p. 125
“The clone fss6 producing red-brown pigment was isolated and further characterized in the study. Based on the physico-chemical characteristics of the pigment, the red-brown pigment was identified as melanin.” - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melaninin Escherichia coli p. 129
"[...] the production of melanin was correlated with homogentistic acid (HGA). The p-hydroxyphenylpyruvate produced by the Escherichia coli host was converted to HGA through the oxidation reaction of introduced HPPD." - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melaninin Escherichia coli p. 124
"HPLC analysis for HGA from the cell-free filtrate of cultures showed that HGA was produced by recombinant E.coli strains before melanin was produced....HGA is the product of ORF2 and should be the precursor of melanin." - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melaninin Escherichia coli p. 129
"The cloned gene (ORF2) encoding putative 4-hydroxyphenylpyruvate dioxygenase (HPPD) was responsible for the production of melanin in E. coli." - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melaninin Escherichia coli p. 129
"The known pathway for the catabolism of tyrosine to HGA in mammals and bacteria is that tyrosine is converted to p-hydroxyphenylpyruvate (p-HPP) through a transamination reaction. Then, p-HPP is converted to HGA through the oxidation of HPPD (Lindstedt and Odelhög 1987; Lindstedt et al. 1977). In the study, the transaminase for the initial step should exist in clone fss6. Three aminotransferase capable of converting tyrosine into p-HPP have been identified in E. coli (Pittard 1987). We propose that tyrosine is first catabolized by aminotransferases of E. coli host, then, the produced p-HPP is converted to HGA by the introduced HPPD gene products. Lastly, the accumulation and polymerization of HGA results in the formation of melanin." - Characterization of a Deep-Sea Sediment Metagenomic Clone that Produces Water-Soluble Melaninin Escherichia coli p. 130
MITF
Catalase
Superoxide dismutase
Mast cell growth factor (MGF)
Basic fibroblast growth factor (bFGF)
Vitamin D
Vitamin D has shown to be beneficial for vitiligo patients. Vitamin D would be secreted from the bacteria, penetrating the skin, and enter the melanocytes.
“Furthermore, vitamin D and its chemically engineered analogs are supposed of being involved in the control of multiple intracellular pathways responsible for the melanin synthesis and melanocyte survival and in the suppression of the immune response. All these findings encouraged clinicians to further investigate the effects of treatment with vitamin D compounds in vitiligo patients, thus opening new perspectives in the therapy of depigmentation.” - New Insights on Therapy with Vitamin D Analogs Targeting the Intracellular Pathways That Control Repigmentation in Human Vitiligo, pp. 18-19
“Vitamin D ligands control multiple critical biological functions and play different roles depending on the type of the tissue targeted and on the mechanism(s) involved in the cellular signaling. These compounds, acting mainly through their nuclear receptors (vitamin D receptor—VDR), protect the epidermal melanin unit and restore the melanocyte integrity by two main mechanisms (reviewed in Birlea et al.33): controlling the activation, proliferation, and migration of melanocytes and the pigmentation pathways, and modulating the T cell activation that is apparently correlated with the melanocyte disappearance in vitiligo. The multiple effects of VDR on immune cells lead to the recognition of vitamin D as a potent immunomodulator.” - New Insights on Therapy with Vitamin D Analogs Targeting the Intracellular Pathways That Control Repigmentation in Human Vitiligo, p.19
“Experiments using normal melanocytes62,63 and melanoma cells64 of animal and human origin showed a stimulatory effect of vitamin D compounds on melanocyte differentiation and tyrosinase activity. Studies showed that topical application of vitamin D3 on the epidermis of mice increased the number of DOPA-positive melanocytes.65 Indirect data about the stimulatory effect of vitamin D on tyrosinase were provided by animal models fed with vitamin D, which in contrast with vitamin D-deficient animals showed a greater skin tyrosinase activity in response to UV irradiation.66” - New Insights on Therapy with Vitamin D Analogs Targeting the Intracellular Pathways That Control Repigmentation in Human Vitiligo, p.19
(“Opposed to its stimulatory effects on melanocyte differentiation, vitamin D was also reported to have an inhibitory effect on the melanocyte growth,67–69 an effect that was confirmed on several other cell types as well. As such, vitamin D has been shown to act as a regulator of replication in keratinocytes, and has been reported to coordinate the growth arrest in several types of cancer cells such as prostatic, breast, colon, leukemic, squamous carcinoma, melanoma, and Kaposi sarcoma cells.56,70,71 It may be inferred that while the stimulatory effect of vitamin D compounds on tyrosinase activity and the modulatory effect on T cell infiltrate may enhance repigmentation in vitiligo, their suppressor effect on melano- cyte growth would minimize or delay the pigment restoration. Therefore, designing new vitamin D analogs able to stimulate melanocyte differentiation and to enhance tyrosinase activity is of a particularly high interest in vitiligo.
The mechanism through which vitamin D exerts its effects on melanocytes is not yet fully understood. It is believed that vitamin D is involved in melanocyte physiology by coordinating the melanogenic cytokines (most likely endothelin-3 (ET-3)) and the activity of SCF/c-Kit system—one of the most important regulators of melanocyte viability and maturation.33” - New Insights on Therapy with Vitamin D Analogs Targeting the Intracellular Pathways That Control Repigmentation in Human Vitiligo, p.20)
“Furthermore, a proposed mechanism involving vitamin D in the protection of vitiliginous skin is based on its antioxidant properties and regulatory function toward the ROS that are produced in excess in vitiligo epidermis. It is also believed that vitamin D coordinates calcium fluxes in melanocytes, a similar function to that exerted in other cell types such as human keratinocytes, hepatocytes, and murine B16 melanoma cells.33,72,73 Based on these observations, it has been suggested that vitamin D is involved in the correction of the aberrant calcium fluxes that accompany the melanocytic loss in vitiligo.33,34,74,75 Several other studies emphasized the complex anti-apoptotic action of vitamin D on melanocytes76 and keratinocytes.77,78 It is likely that vitamin D can protect pigment cells against apoptosis, one of the cytotoxic mechanisms proposed among those causing melanocyte disappearance in vitiligo.” - New Insights on Therapy with Vitamin D Analogs Targeting the Intracellular Pathways That Control Repigmentation in Human Vitiligo, p.20
Andreas: Keep in mind that vitamin D (in the human body at least) is produced from a precursor that requires (UV?) light to be converted into vitamin D. Maybe this does not apply to bacteria, I don’t know.
Nina: Size of vit D: ~400 g/mol
Vitamin B12 (cyanoocobalamin)
“The biosynthesis pathway has been extensively characterized in several lactic acid bacteria, including Lactobacillus plantarum WCFS1” - High-Level Folate Production in Fermented Foods by the B12 Producer Lactobacillus reuteri JCM1112
"The constitutive overexpression of the folate biosynthesis genes of L. plantarum WCFS1 in cultures of L. reuteri JCM1112/pNZ7026 resulted in an almost 100-fold increase in folate levels, while the control (L.reuteri JCM1112/pNZ7021) did not show any change in folate and B12 production. The overproduction of folate was found to have a very small effect on B12 production (10% reduction), resulting in a folate/B12 ratio of approximately 100:1 (wt/wt)" - High-Level Folate Production in Fermented Foods by the B12 Producer Lactobacillus reuteri JCM1112
“The same construct has been tested with L. plantarum, resulting in similar folate production levels (28), and similar results were obtained when the same strategy was applied to Lactococcus lactis (30) and Lactobacillus gasseri (29).” - High-Level Folate Production in Fermented Foods by the B12 Producer Lactobacillus reuteri JCM1112
“In this study, we demonstrated that it is possible to combine the production of folate and the production of B12 in L. reute” - The complete coenzyme B12 biosynthesis gene cluster of Lactobacillus reuteri CRL1098
“Vitamin B12 consists of a tetrapyrrolic-derived corrin ring with a cobalt ion chelated at the core” - The complete coenzyme B12 biosynthesis gene cluster of Lactobacillus reuteri CRL1098
“Coenzyme B12 biosynthesis is limited to a few representatives of bacteria and archaea (Martens et al., 2002). It appears that B12-dependent enzymes are absent from plants and fungi, but widespread in prokaryotes, protists and animals” - The complete coenzyme B12 biosynthesis gene cluster of Lactobacillus reuteri CRL1098
“Lb. reuteri was the first lactic acid bacterium reported to be able to produce B12” - The complete coenzyme B12 biosynthesis gene cluster of Lactobacillus reuteri CRL1098
“In this study, we extend the analysis of the presumed coenzyme B12 biosynthesis gene cluster of Lb. reuteri and describe the presence of a complete gene cluster encoding all the enzymic machinery necessary for the de novo synthesis of this important cofactor.” - The complete coenzyme B12 biosynthesis gene cluster of Lactobacillus reuteri CRL1098
Nina: Size of vit B: ~440/1300 g/mol
 "
"