Team:Stockholm/18 July 2010
From 2010.igem.org
(→Andreas) |
(→Colony PCR of pEX constructs) |
||
| Line 16: | Line 16: | ||
''Continued from 17/7'' | ''Continued from 17/7'' | ||
| - | A new colony PCR was run on the same clones as were picked 17/7, using the same tube settings except (1) 1.5 μl primer was used instead of 1.0 μl and (2) 1 μl cell suspension was used instead of 0.5 μl; dH<sub>2</sub>O volume was decreased accordingly. Samples pEX.SOD A and pEX.SOD B were | + | A new colony PCR was run on the same clones as were picked 17/7, using the same tube settings except (1) 1.5 μl primer was used instead of 1.0 μl and (2) 1 μl cell suspension was used instead of 0.5 μl; dH<sub>2</sub>O volume was decreased accordingly. Samples pEX.SOD A and pEX.SOD B were omitted. |
'''PCR settings''' | '''PCR settings''' | ||
Revision as of 20:56, 29 July 2010
Contents |
Andreas
BL21 restreaks
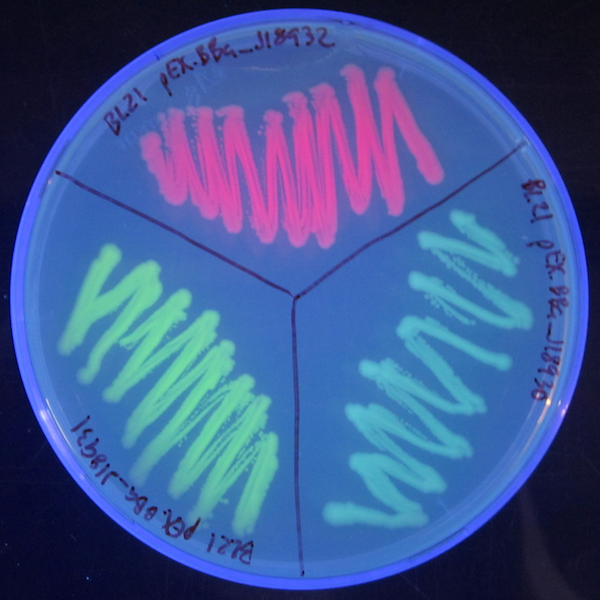
Results from 17/7 restreaks
Nice fluorescent cell colonies appeared on the plate, indicating that IPTG induction from the pEX vector indeed works and that I have successfully inserted the three fluorescent protein genes into pEX.
Competent Top10
Continued from 17/7 At 9:30 AM cell culture still hadn't reached OD600 0.3, only 0.02. I therefore transfered the cells to 37°C, 250 rpm until an OD600 of 0.3. I then followed our standard Top10 protocol.
Colony PCR of pEX constructs
Continued from 17/7
A new colony PCR was run on the same clones as were picked 17/7, using the same tube settings except (1) 1.5 μl primer was used instead of 1.0 μl and (2) 1 μl cell suspension was used instead of 0.5 μl; dH2O volume was decreased accordingly. Samples pEX.SOD A and pEX.SOD B were omitted.
PCR settings
- 95°C - 5:00
- 95°C - 1:00
- 60°C - 0:30
- 72°C - 1:15 (Cycle to step 2. 29 times)
- 72°C - 5:00
- 15°C - ∞
Gel verification
1 % agarose, 90 V, 40 min
Results
Not all bands present in all lanes. Due to lack of time, all samples were discarded and I will pick/verify new colonies after my vacation.
 "
"