Team:Newcastle/12 July 2010
From 2010.igem.org
(→Streak Plates) |
|||
| Line 43: | Line 43: | ||
{| | {| | ||
|- | |- | ||
| - | |[[Image:P7120427.JPG|thumb]] | + | |[[Image:P7120427.JPG|thumb]right] |
|[[Image:P7120453.JPG|thumb]] | |[[Image:P7120453.JPG|thumb]] | ||
|[[Image:P7120450.JPG|thumb]] | |[[Image:P7120450.JPG|thumb]] | ||
Revision as of 15:36, 21 July 2010
Monday
Contents |
Aims
The aim of todays Lab session was to perform the following(see summary) to isolate lacI.
Summary:
- Colony PCR
- Streak plate
- Miniprep PCR- of colonies that worked
Equipment
- PCR reagents
- Agar plates
- Gloves
- Wire loop
Colony PCR
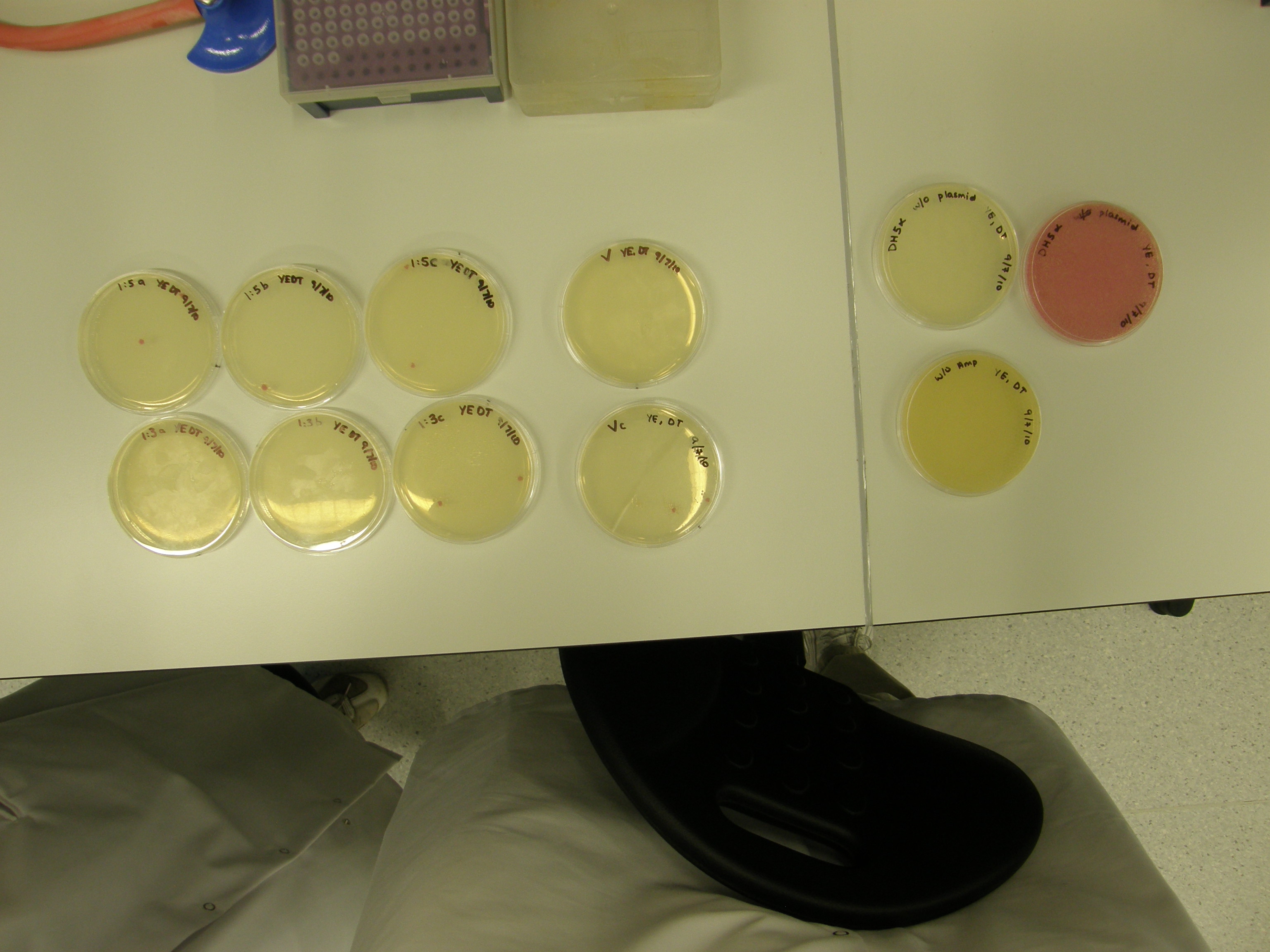
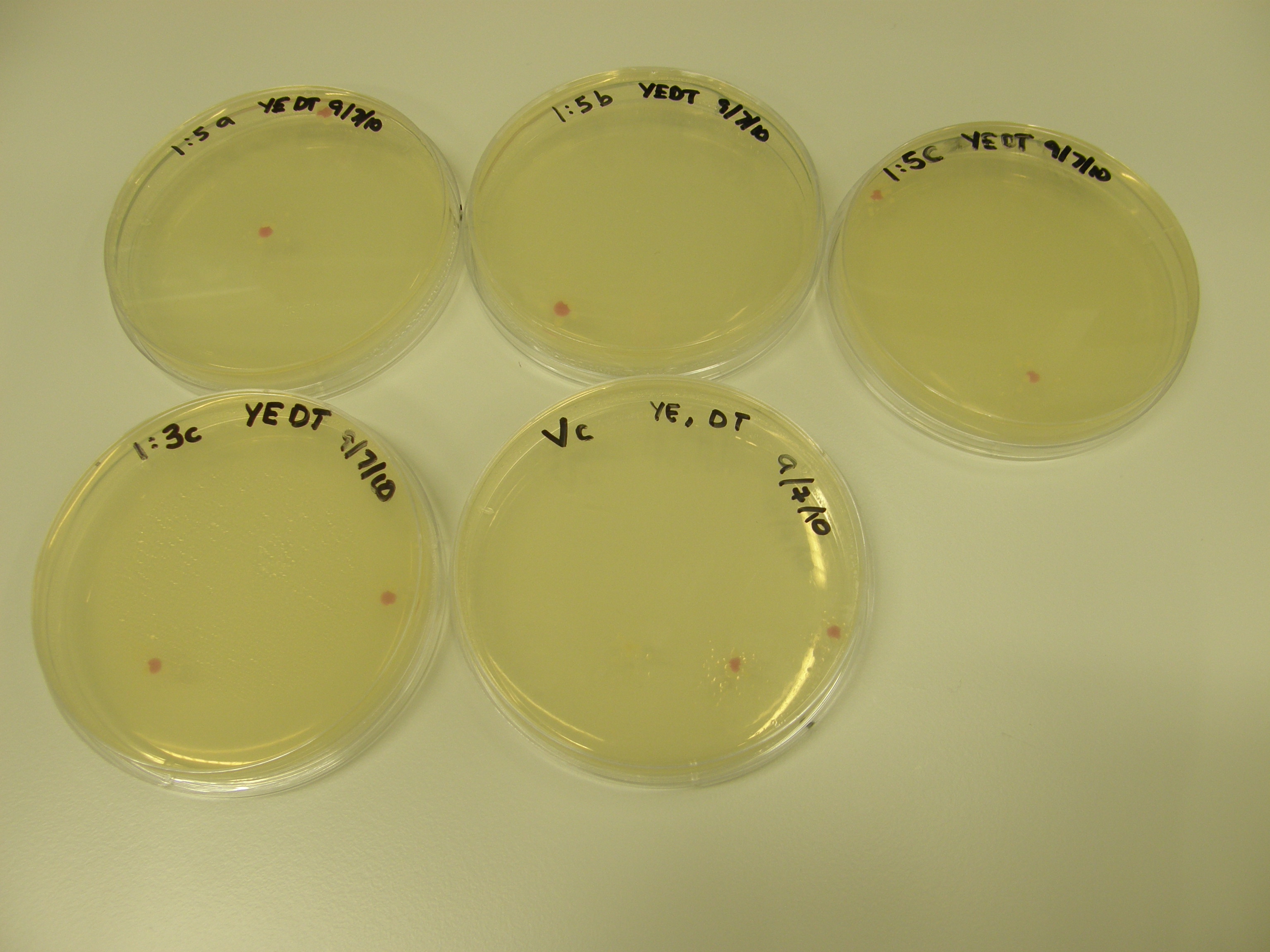
Of the 11 plates with colonies grown 7 were used for colony PCR and Streak plating. Colony PCR is less accurate than the mini prep however results can be seen quicker whereas the miniprep has a 1 day wait. The colonies selected selected were satellite colonies because ampicillin degraded???
7 Tubes were labelled 1-7 (+ a control)
The quantities for the PCR are as follows:
- 1microlitre Template
- 0.25 microlitre GoTaq Polymerase
- 2.5 microlitre forward primer
- 2.5 microlitre reverse primer
- 1 microlitre dNTP (10molar stock)
- 10.0 microlitre 5times GoTaq buffer
- 32.75 microlitre deionised H20
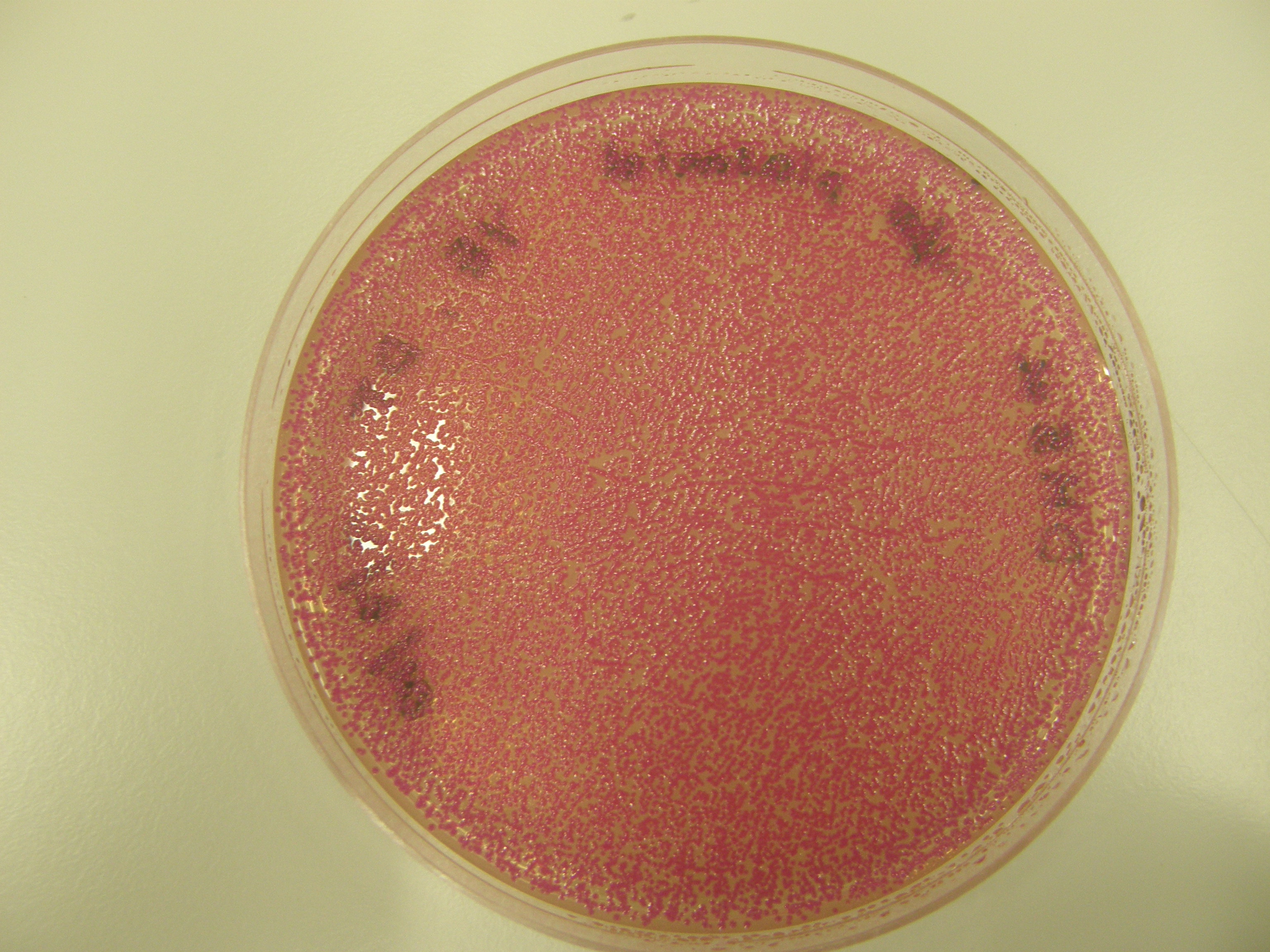
Note There are 2 possible reasons for red colonies formed:
- Partial digest- rejoins without insert
- with insert
We need to tell the difference between the two.
Note PCR is best when everything is kept on ice!
| [[Image:P7120427.JPG|thumb]right] |
Note Melting temperatures of primers is important and can have a big effect on the product formed (Temperature used see Thursday)
Spread Plates
Tommorrow
We are going to take one of the colonies that have worked and possible a whole plasmid prep.
 "
"