Team:Cambridge/HIV Diagnosis
From 2010.igem.org
| Line 1: | Line 1: | ||
{{:Team:Cambridge/Templates/header}} | {{:Team:Cambridge/Templates/header}} | ||
| - | + | =HIV Diagnosis= | |
| - | + | One of the projects that the Cambridge team considered was to replace the [http://en.wikipedia.org/wiki/ELISA ELISA] currently used for HIV diagnosis with a single celled biosensor. This would have a number of advantages: | |
| - | ** | + | *Cheap, simply taking a sample from a fermenter |
| - | ** | + | *Easy to interpret - pigment production as a digital signal, this would allow a member of the community to administer the test rather than it being sent off to a lab |
| - | ** | + | HIV was chosen because diagnosis alone is useful for this disease in a way in which it is not for many others. |
| - | * | + | ==Methods== |
| - | + | Two broad methods were considered, both of which relied on detecting antibodies in the patient's serum. | |
| - | + | ===RTKs=== | |
| + | Receptor tyrosine kinases which would phosphorylate only when brought close together, with external domain presenting antigens which would be bridged by antigens | ||
| + | ===Tethering approach=== | ||
| + | [[Image:Hiv.png|680px]] | ||
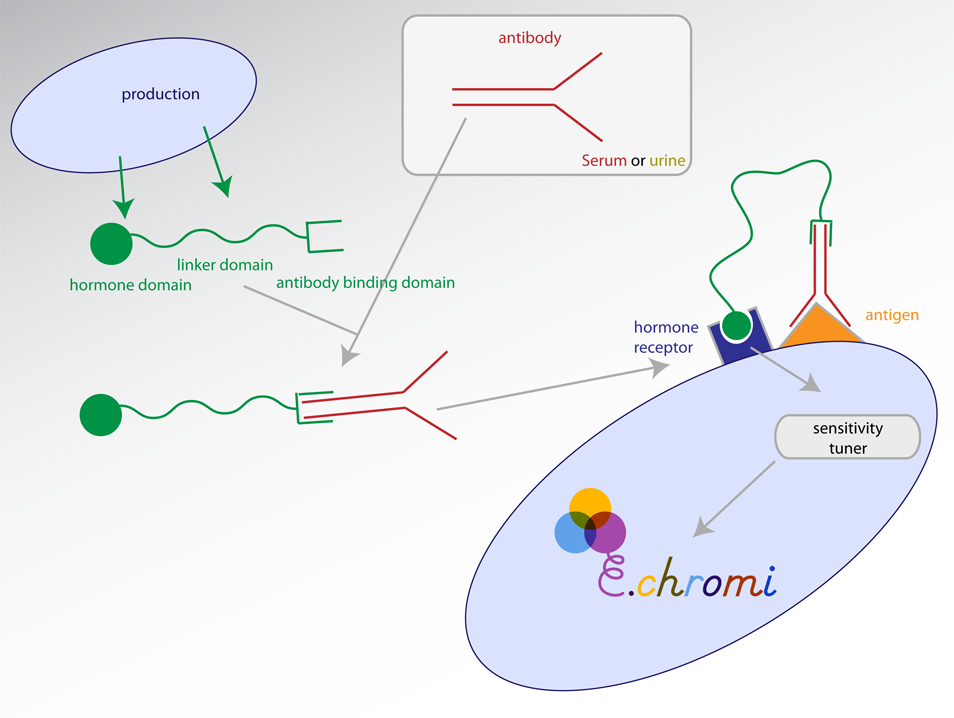
| + | *A fusion protein would be produced comprising | ||
| + | **A general antibody binding domain e.g. Staphylococcus protein A (which has been biobricked) | ||
| + | **A linker domain | ||
| + | **A peptide signal of some description, (which has a characterised detector) | ||
| + | *This would be detected by cells expressing on their surface both: | ||
| + | **The antigen - which would localise antibodies (with bound fusion proteins) close to the cell | ||
| + | **The peptide signal detector - which would detect this localisation | ||
| + | *Activation of the peptide signal receptor would trigger the production of a coloured pigment output via a sensitivity tuner. | ||
| + | |||
| + | The most challenging aspect was finding an appropriate large peptide and receptor. The most promising was a yeast mating hormone (used in yeast) but we concluded that the processing of this small peptide would likely by affected by it's fusion to other hormones and abandoned the idea. If this could be overcome we believe the project might have a lot of potential | ||
{{:Team:Cambridge/Templates/footer}} | {{:Team:Cambridge/Templates/footer}} | ||
Revision as of 16:06, 17 July 2010

HIV Diagnosis
One of the projects that the Cambridge team considered was to replace the [http://en.wikipedia.org/wiki/ELISA ELISA] currently used for HIV diagnosis with a single celled biosensor. This would have a number of advantages:
- Cheap, simply taking a sample from a fermenter
- Easy to interpret - pigment production as a digital signal, this would allow a member of the community to administer the test rather than it being sent off to a lab
HIV was chosen because diagnosis alone is useful for this disease in a way in which it is not for many others.
Methods
Two broad methods were considered, both of which relied on detecting antibodies in the patient's serum.
RTKs
Receptor tyrosine kinases which would phosphorylate only when brought close together, with external domain presenting antigens which would be bridged by antigens
Tethering approach
- A fusion protein would be produced comprising
- A general antibody binding domain e.g. Staphylococcus protein A (which has been biobricked)
- A linker domain
- A peptide signal of some description, (which has a characterised detector)
- This would be detected by cells expressing on their surface both:
- The antigen - which would localise antibodies (with bound fusion proteins) close to the cell
- The peptide signal detector - which would detect this localisation
- Activation of the peptide signal receptor would trigger the production of a coloured pigment output via a sensitivity tuner.
The most challenging aspect was finding an appropriate large peptide and receptor. The most promising was a yeast mating hormone (used in yeast) but we concluded that the processing of this small peptide would likely by affected by it's fusion to other hormones and abandoned the idea. If this could be overcome we believe the project might have a lot of potential
 "
"