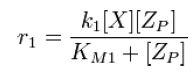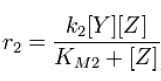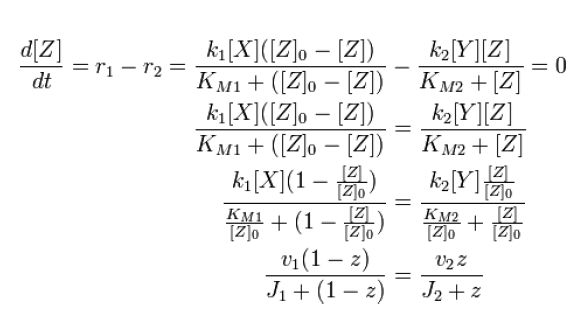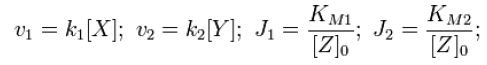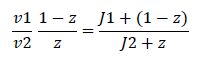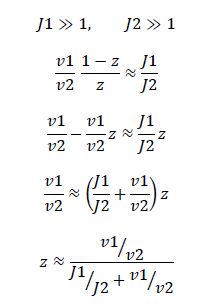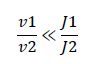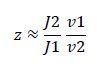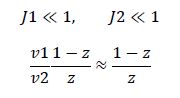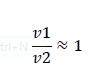Team:Stanford/Research/Modeling
From 2010.igem.org
Drummstikk (Talk | contribs) (→Kinase/Phosphatase System) |
Drummstikk (Talk | contribs) (→Kinase/Phosphatase System) |
||
| Line 451: | Line 451: | ||
From Michaelis-Menten kinetics we know that the rate at which Zp is dephosphorylated is | From Michaelis-Menten kinetics we know that the rate at which Zp is dephosphorylated is | ||
| - | [[Image:R1.JPG]][[Team:Stanford/Research/Modeling | + | [[Image:R1.JPG]][[Team:Stanford/Research/Modeling#References| [2]]] |
and the rate at which Z is phosphorylated is | and the rate at which Z is phosphorylated is | ||
| - | [[Image:R2.JPG]][[Team:Stanford/Research/Modeling | + | [[Image:R2.JPG]][[Team:Stanford/Research/Modeling#References| [2]]] |
We also know that Z can be modified an unmodified, but its total is conserved: [Z]o = [Zp] + [Z] | We also know that Z can be modified an unmodified, but its total is conserved: [Z]o = [Zp] + [Z] | ||
| Line 461: | Line 461: | ||
At steady state, modification and unmodification rates are equal, leading us to | At steady state, modification and unmodification rates are equal, leading us to | ||
| - | [[Image:1stblock.JPG]][[Team:Stanford/Research/Modeling | + | [[Image:1stblock.JPG]][[Team:Stanford/Research/Modeling#References| [2]]] |
where | where | ||
| - | [[Image:2nd_block.JPG]][Team:Stanford/Research/Modeling | + | [[Image:2nd_block.JPG]][[Team:Stanford/Research/Modeling#References| [2]]] |
Revision as of 02:24, 28 October 2010

| Home | Project | Applications | Modeling | Parts | Team | Notebook |
Contents |
Goals
Our intuition for what makes a good ratio sensor could only take us so far. From the very first stages of design, we wanted to back up and test our ideas with mathematical tools. Luckily, we found that solving the equations of mass action kinetics at steady-state was enough to give us clear design criteria. We present the mathematical basis for sensors that are capable of sensing a single ratio digitally, or many ratios in an analog fashion.
The System
sRNA System
Boolean model for the sRNA system. This is mean to provide a significantly simplified mathematical understanding of the dynamics involved.
clear all clc %Constants and Values trackingNum = 6; %mRNA_pA, mRNA_pB, sRNA_A, sRNA_B, protein_A, protein_B; inputNum = 2; %A, B; processNum = 6; %Trx_pA, Trx_sRNA_A, Trx_pB, Trx_sRNA_B, Trl_A, Trl_B; %Cell/Matrix Dimensions m = 2^trackingNum; n = 2^inputNum; c = cell(m+1, n+1); trackingMatrix = de2bi(0:m-1); inputMatrix = de2bi(0:n-1); processMatrix = zeros(1,processNum); %Populating the Cell (Tracking x Input) c{1,1} = [0]; for j = 1 for i = 2:m+1 c{i,j} = trackingMatrix(i-1,:); end end for i = 1 for j = 2:n+1 c{i,j} = inputMatrix(j-1,:); end end for i = 2:m+1 for j = 2:n+1 c{i,j} = processMatrix; end end %Rules, Acessing, and Changing for j = 2:n+1 A = c{1,j}(1); B = c{1,j}(2); for i = 2:m+1 mRNA_pA = c{i,1}(1); mRNA_pB = c{i,1}(2); sRNA_A = c{i,1}(3); sRNA_B = c{i,1}(4); protein_A = c{i,1}(5); protein_B = c{i,1}(6); %Trx_pA if A == 1 c{i,j}(1) = 1; end %Trx_pB if B == 1 c{i,j}(2) = 1; end %Trx_sRNA_A if A == 1 c{i,j}(3) = 1; end %Trx_sRNA_B if B == 1 c{i,j}(4) = 1; end %Trl_A if mRNA_pA == 1 && mRNA_pB == 0 c{i,j}(5) = 1; end %Trl_B if mRNA_pB == 1 && mRNA_pA == 0 c{i,j}(6) = 1; end end end %Display Results (Column by Column) [nrows,ncols]= size(c); %Condense Cell and Display for i = 1:nrows for j = 1:ncols string = num2str(c{i,j}); l = length(string); r = 1; s = 1; t = 0; while t ~= 1 if r == l t = 1; end noSpacesString(s) = string(r); r = r+3; s = s+1; end c{i,j} = noSpacesString; end end c(:,:) %Find Steady States and Corresponding Inputs counter = 1; for i = 2:m+1 for j = 2:n+1 if c{i,j} == c{i,1} completeMatrix{counter,1} = c{i,j}; %Steady state values completeMatrix{counter,2} = num2str([i-1,j-1]); %Location (n x m) within results area completeMatrix{counter,3} = c{1,j}; %Corresponding input values counter = counter+1; end end end SS_mXn_Input = completeMatrix time = 0:processNum; %plot(time,,time,,time,,time,,time,,time,) %New Cell for Specific Cases % d(:,1) = c(:,1); % for j = 2:n+1 % C1 = '0000'; % C2 = '0011'; % C3 = '0100'; % C4 = '0111'; % C5 = '1000'; % C6 = '1011'; % C7 = '1100'; % % a = {C1, C2, C3, C4, C5, C6, C7}; % for b = 1:1:length(a) % if strcmp(c(1,j),a(b)) == 1 % d(:,b+1) = c(:,j); % end % end % end % d
ans =
'0' '00' '01' '10' '11'
'000000' '000000' '010100' '101000' '111100'
'000001' '000000' '010100' '101000' '111100'
'000010' '000000' '010100' '101000' '111100'
'000011' '000000' '010100' '101000' '111100'
'000100' '000000' '010100' '101000' '111100'
'000101' '000000' '010100' '101000' '111100'
'000110' '000000' '010100' '101000' '111100'
'000111' '000000' '010100' '101000' '111100'
'001000' '000000' '010100' '101000' '111100'
'001001' '000000' '010100' '101000' '111100'
'001010' '000000' '010100' '101000' '111100'
'001011' '000000' '010100' '101000' '111100'
'001100' '000000' '010100' '101000' '111100'
'001101' '000000' '010100' '101000' '111100'
'001110' '000000' '010100' '101000' '111100'
'001111' '000000' '010100' '101000' '111100'
'010000' '000001' '010101' '101001' '111101'
'010001' '000001' '010101' '101001' '111101'
'010010' '000001' '010101' '101001' '111101'
'010011' '000001' '010101' '101001' '111101'
'010100' '000001' '010101' '101001' '111101'
'010101' '000001' '010101' '101001' '111101'
'010110' '000001' '010101' '101001' '111101'
'010111' '000001' '010101' '101001' '111101'
'011000' '000001' '010101' '101001' '111101'
'011001' '000001' '010101' '101001' '111101'
'011010' '000001' '010101' '101001' '111101'
'011011' '000001' '010101' '101001' '111101'
'011100' '000001' '010101' '101001' '111101'
'011101' '000001' '010101' '101001' '111101'
'011110' '000001' '010101' '101001' '111101'
'011111' '000001' '010101' '101001' '111101'
'100000' '000010' '010110' '101010' '111110'
'100001' '000010' '010110' '101010' '111110'
'100010' '000010' '010110' '101010' '111110'
'100011' '000010' '010110' '101010' '111110'
'100100' '000010' '010110' '101010' '111110'
'100101' '000010' '010110' '101010' '111110'
'100110' '000010' '010110' '101010' '111110'
'100111' '000010' '010110' '101010' '111110'
'101000' '000010' '010110' '101010' '111110'
'101001' '000010' '010110' '101010' '111110'
'101010' '000010' '010110' '101010' '111110'
'101011' '000010' '010110' '101010' '111110'
'101100' '000010' '010110' '101010' '111110'
'101101' '000010' '010110' '101010' '111110'
'101110' '000010' '010110' '101010' '111110'
'101111' '000010' '010110' '101010' '111110'
'110000' '000000' '010100' '101000' '111100'
'110001' '000000' '010100' '101000' '111100'
'110010' '000000' '010100' '101000' '111100'
'110011' '000000' '010100' '101000' '111100'
'110100' '000000' '010100' '101000' '111100'
'110101' '000000' '010100' '101000' '111100'
'110110' '000000' '010100' '101000' '111100'
'110111' '000000' '010100' '101000' '111100'
'111000' '000000' '010100' '101000' '111100'
'111001' '000000' '010100' '101000' '111100'
'111010' '000000' '010100' '101000' '111100'
'111011' '000000' '010100' '101000' '111100'
'111100' '000000' '010100' '101000' '111100'
'111101' '000000' '010100' '101000' '111100'
'111110' '000000' '010100' '101000' '111100'
'111111' '000000' '010100' '101000' '111100'
SS_mXn_Input =
'000000' '1 1' '00'
'010101' '22 2' '01'
'101010' '43 3' '10'
'111100' '61 4' '11'
Kinase/Phosphatase System
Throughout this derivation, we refer to Goldbeter and Koshland's seminal work on the kinetics of emzyme pairs [1].
For an opposing pair of enzymes X and Y modifying and unmodifying a substrate Z with modification p,
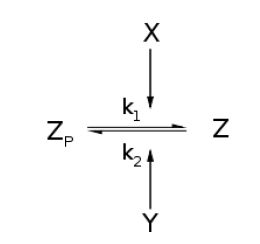
From Michaelis-Menten kinetics we know that the rate at which Zp is dephosphorylated is
and the rate at which Z is phosphorylated is
We also know that Z can be modified an unmodified, but its total is conserved: [Z]o = [Zp] + [Z]
At steady state, modification and unmodification rates are equal, leading us to
where
Rearranging gives
Now, assume that the two opposing enzymes are nowhere near being saturated with substrate:
So, the fraction of modified substrate is a saturating function of the ratio of inputs. The half-max value is the ratio of the strengths of the two enzymes. The linear regime of this function exists where
and the fraction of unmodified substrate is a linear function of the ratio of inputs.
We now know that the requirements for this analog ratio sensor are
- Consistent relationships between inputs and enzyme activities (non-trivial)
- A non-saturating amount of substrate
But what happens if we disobey requirement 1? Let's increase the amount of substrate to saturation so that
When the enzymes are saturated, the only steady-state solution is v1 = v2. Any ratio of v1/v2 below this, and all the substrate becomes modified. Any ratio above and all the substrate becomes unmodified. Simply by changing the concentration of substrate, we have converted our analog ratio sensor to a digital ratio sensor.
The requirements for this digital ratio sensor are:
- Consistent relationships between inputs and enzyme activities (non-trivial)
- An amount of substrate sufficient to saturate both opposing enzymes
References
[1] [http://www.ncbi.nlm.nih.gov/pubmed/6947258 Goldbeter A, Koshland DE Jr.: An amplified sensitivity arising from covalent modification in biological systems. PNAS. 1981 Nov;78(11)6840-4.]
[2] [http://en.wikipedia.org/wiki/Goldbeter-Koshland_kinetics Goldbeter-Koshland kinetics In Wikipedia. Retrieved July 13, 2010, from http://en.wikipedia.org/wiki/Goldbeter-Koshland_kinetics]
 "
"