Team:Wisconsin-Madison/results
From 2010.igem.org
(→Conclusion) |
|||
| Line 67: | Line 67: | ||
====Results==== | ====Results==== | ||
| - | + | [[Image:UW L Fucose.jpg|400px|center]] | |
| + | Figure 2: L-Fucose content of plasmids K318500, K318501, and K200021 (control) in backbone pSB1AK3 transformed in MG1655 | ||
| + | |||
| + | ====Conclusion==== | ||
| + | We assume the amount of Colonic Acid produced is linearly related L-Fucose content. This graph shows the amount of L-Fucose found in cultures containing our induced constructs. We found the amount of Colonic Acid produced in the samples with RcsB being over expressed showed significantly increased production over the control cells. More samples must be assayed to tighten the error bars to make the amount produced by the cells with RcsA being overexpressed statistically significant. Unfortunately, our cells containing the plasmid that produces a combination of RcsA and RcB would not grow strong enough for testing. We hope to add this sample to our data set soon. | ||
<br> | <br> | ||
Revision as of 02:23, 28 October 2010
Encapsulation
| Part Number | Function | Expression Type | Zip File |
| <partinfo>BBa_k318500</partinfo> | Produces Trascription Factor RcsA | Inducible - IPTG | 500 |
| <partinfo>BBa_k318501</partinfo> | Produces Trascription Factor RcsB | Inducible - IPTG | 501 |
| <partinfo>BBa_k318502</partinfo> | Produces Trascription Factor RcsA & RcsB | Inducible - IPTG | 502 |
| <partinfo>BBa_k200021</partinfo> | Empty Vector/Contol | Inducible - IPTG | NA |
Our goals is to have each cell be surrounded by a protective 'capsule' to allow them to safely travel through the harsh acidic environment of the stomach to arrive in the small intestine for their main purpose. Imperial 2009 used Transcription Factor RcsB to stimulate a the production of Colonic Acid from the capsule synthesis pathway of E.coli. Colonic Acid s a polysaccharide containing a repeat unit with D-glucose, L-fucose, D-galactose, and D-glucuronate. Colonic Acid has been shown to increase cell survivability in acidic conditions. RcsA and RcsB are transcription factors that are know to be positive regulators of capsular polysaccharides synthesis. We placed RcsA, RcsB, and a combination of the two under a IPTG inducible promoter to test both quantity of colonic acid produced and cell survivability. These two transcription factors form a heterodimer that is know to activate around 19 genes related to colonic acid synthesis. RcsB is also know to form a homodimmer and positively regulate cell division. RcsA and RcsB belong to the multicomponent RcsF/RcsC/RcsD/RcsA-RcsB phosphorelay system.
Colonic Acid Quantification
Background
Colonic Acid is a polysaccharide containing a repeat unit with D-glucose, L-fucose, D-galactose, and D-glucuronate. Biological extracts often contain compounds, which under heating with H2SO4 yield brown products absorbing between 396 nm and 427 nm. Colonic acid can be estimated by measuring L-fucose content.
Download procedure here
Results
Figure 2: L-Fucose content of plasmids K318500, K318501, and K200021 (control) in backbone pSB1AK3 transformed in MG1655
Conclusion
We assume the amount of Colonic Acid produced is linearly related L-Fucose content. This graph shows the amount of L-Fucose found in cultures containing our induced constructs. We found the amount of Colonic Acid produced in the samples with RcsB being over expressed showed significantly increased production over the control cells. More samples must be assayed to tighten the error bars to make the amount produced by the cells with RcsA being overexpressed statistically significant. Unfortunately, our cells containing the plasmid that produces a combination of RcsA and RcB would not grow strong enough for testing. We hope to add this sample to our data set soon.
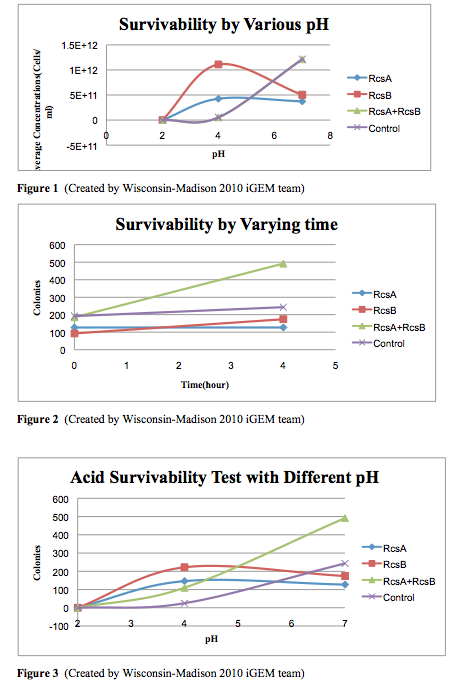
Cell Survivability in Low pH
Background
Download procedure here
Results
Conclusion
The cells survivability testing is done in three different pH conditions, pH=2, 4, and 7, for 3 samples and one controls. We use identical cells, which is MG1655, to generate the experimental data. After four hour exposure in the acidic medium, we observed that there are no survivals under the pH=2 condition. However, there is an significant improvement at pH 4 in the experimental strains. Moreover, from the experiments, we observed that the combination of having RcsA and RcsB both presented enhanced the survivability of the experimental straints. Our result from this testing quantified can confirmed the cells survivability at low pH levels.
Timed Lysis
| Part Number | Function | Induction | Zip File |
| <partinfo>BBa_k318513</partinfo> | Produces RFP | stationary phase | 513 |
Background
gadAp: A pH Sensitive Promoter Based on available literature (see Project Page), we expected an increase in expression from gadAp (BBa_K318512) under both acidic pH conditions (from pH 5 to 3) and in the stationary phase (after an OD of 1 or 2). As most previous studies of gadAp had focused on gadA mRNA production, we wanted to instead explore the viability of gadAp for use in expression of protein. Given the stressful conditions under which it acts and its complex regulation, the possibility that mRNA-level regulation or an inability of acid-stressed cells to produce large quantities of protein may exist could not be discounted. As such, we combined the gadA promoter with a RFP gene (BBa_K318513) to directly test its ability to produce protein under stationary phase and acid stress conditions.
Low pH/Stationary Phase Induction
Download testing procedure (.pdf) here.
Download example spreadsheet (.xls) here.
Below are two figures of sample data:
Figure 4. Lines represent OD600 and are associated with the right axis. Dots indicate RFU/OD600 and associated with the left axis. In this trial, maximum induction occurred at an OD600 of approximately 6 (right axis, associated with line) and yielded about 10000 RFU/OD. Note that immediately before maximum OD600 was reached, RFU/OD increased about 5-fold. Controls included for this trial were a lacI promoter + RBS + mRFP1 + TT construct in the same plasmid (pSB1A2) and a WT control of MG1655 cells. These controls and results indicate that (1) mRFP1 expression by gadAp at high OD600 is a function of the gadA promoter, not of promoters at large (lacI promoter did not express) or of the cells themselves (MG1655 did not fluoresce).
Figure 5.This trial was run parallel to the pH 7 trial in Figure 4. It indicates that cells grown in LB media at pH 5.5 do not exhibit a drastically different promotion behavior from their gadA promoters than those grown in neutral media.
Conclusion
We have shown that in MG1655 grown in LB media, gadAp is strongly induced by entry into the final stages (where increase in OD completely halts) of cell culture growth. In addition, it is apparent that the promoter has very tight (no leaky expression) control of expression during the exponential and early stationary phase. However, despite supporting the literature and our expectations in terms of stationary phase induction, use of this construct is inappropriate for the goals of our project--we are most interested in pH sensitive expression. However, more testing is required; it is possible that pH sensitive expression may occur with acid shock of stationary phase cells or many other conditions. Our attempts to test this hypothesis were unsuccessful as cells tended to precipitate out of solution during acid shock.
Encryption
Unfortunately, we never got to testing our systems. We managed to clone a two component encryption system (as detailed here). Rigorous testing of our system never occurred due to time constraints of the competition. Our enzyme delivery took precedence as the competition drew near and our financial resources became more and more strained.
 "
"