Team:Cambridge/Bioluminescence/Firefly Characterisation
From 2010.igem.org
(→Coloured outputs) |
|||
| Line 131: | Line 131: | ||
|} | |} | ||
</center> | </center> | ||
| - | |||
=Compatibility= | =Compatibility= | ||
Revision as of 00:16, 28 October 2010

This page describes characterisation for part [http://partsregistry.org/Part:BBa_K325219 BBa_K325219], Red luciferase and LRE operon from L. cruciata under pBAD. It also gives a comparison of the light ouput of the different coloroured mutants we produced.
- Description
- Arabinose to light
- Luciferin to light
- [[Team:Cambridge/Bioluminescence/Firefly_Characterisation#Effect_of_pH|Effect of pH
- Coloured outputs
Description

This part generates a red-mutant of the luciferase from the Japanese firefly (L.cruciata) as well as the luciferin regenerating enzyme (LRE). It is under the control of an Arabinose induced promoter. D-Luciferin has to be added to obtain light output.
Arabinose to light
This page describes the relationship between Arabinose concentration in the medium with light output. We used a [http://www.bmglabtech.com/products/microplate-reader/instruments.cfm?product_id=2 FLUOstar OPTIMA] microplate reader to quantify the light output. Protocol and plate reader settings used are given below.
Data
Transfer function of <partinfo>K325219</partinfo>.
[http://partsregistry.org/wiki/images/2/26/IntegratedactiPP%2BLRE.png http://partsregistry.org/wiki/images/thumb/c/c3/Luciferineffect.png/569px-Luciferineffect.png]
The data points represent the mean of 11 values obtained for light output at 30 min interval from 450 min to 750 min after injection of D-Luciferin. These values are the mean of 3 readings as shown in Figure 3. The corresponding error bars represent an interval of twice the standard deviation across the 33 data points centred around the mean value.
Maximum luminescence output of <partinfo>K325219</partinfo> as a function of Arabinose concentration.
[http://partsregistry.org/wiki/images/0/05/MaximumlumPP%2BLRE.png http://partsregistry.org/wiki/images/thumb/0/05/MaximumlumPP%2BLRE.png/569px-MaximumlumPP%2BLRE.png]
These values are the mean of 3 readings as shown in Figure 3. The corresponding error bars represent an interval of twice the standard deviation across the 3 data points centred around the mean value.
Evolution of luminescence with time at different Arabinose concentrations.
[http://partsregistry.org/wiki/images/4/4c/TimePP%2BLRE.png http://partsregistry.org/wiki/images/thumb/4/4c/TimePP%2BLRE.png/569px-TimePP%2BLRE.png]
The interval between measurements is 30 min. Mean values and error bars are based on 3 time repeats.
| Data | Notes | Date Uploaded |
|---|---|---|
| [http://partsregistry.org/wiki/images/8/82/BBa_K325219ArabinosetoLight.xls Media:BBa_K325219ArabinosetoLight.xls] | Raw data from experiment | 21/10/2010 |
Protocol
Luciferin to light
The light output is also a function of the concentration of D-Luciferin the media. This page describes the relationship between D-Luciferin concentration and light output. We used a [http://www.bmglabtech.com/products/microplate-reader/instruments.cfm?product_id=2 FLUOstar OPTIMA] microplate reader to quantify the light output. Protocols and plate reader settings used are given below.
Data
Effect of D-Luciferin concentration on light output from <partinfo>K325219</partinfo>.
[http://partsregistry.org/wiki/images/c/c3/Luciferineffect.png http://partsregistry.org/wiki/images/thumb/c/c3/Luciferineffect.png/569px-Luciferineffect.png]
The data points represent the mean of 31 values obtained for light output at 30 min interval from 1500 min to 2400 min after injection of D-Luciferin. These values are the mean of 3 readings as shown in Figure 3. The corresponding error bars represent an interval of twice the standard deviation across the 31 data points centred around the mean value.
Evolution of luminescence with time at different Arabinose concentrations.
[http://partsregistry.org/wiki/images/b/b9/Luctimecourse.png http://partsregistry.org/wiki/images/thumb/b/b9/Luctimecourse.png/569px-Luctimecourse.png]
The interval between measurements is 30 min. Mean values and error bars are based on 3 time repeats.
| Data | Notes | Date Uploaded |
|---|---|---|
| [http://partsregistry.org/wiki/images/9/9a/BBa_K325219luciferintolight.xls Media:BBa_K325219luciferintolight.xls] | Raw data from experiment | 21/10/2010 |
Effect of pH
Both the intensity and the spectrum emitted by the luciferase-luciferin reaction has been shown to be higly dependent on the pH of the medium. The main characterisation experiments have been performed in LB Broth at pH 7, so in order to assess this effect cultures with LB and a citrate buffer were prepared (pH = 5.3, pH 6.1 and pH = 7) This page describes the results of these experiments. We used a [http://www.bmglabtech.com/products/microplate-reader/instruments.cfm?product_id=2 FLUOstar OPTIMA] microplate reader to quantify the light output. Protocols and plate reader settings used are given below.
Data
Maximum light output within 5 hours of D-luciferin injection at different pH values.
[http://partsregistry.org/wiki/images/e/e8/Phhistogram.png http://partsregistry.org/wiki/images/thumb/e/e8/Phhistogram.png/569px-Phhistogram.png]
These values are the mean of 3 readings. The corresponding error bars represent an interval of twice the standard deviation across the 3 data points centred around the mean value.
Evolution of light output at different values of pH.
[http://partsregistry.org/wiki/images/d/d8/Phtimecourse.png http://partsregistry.org/wiki/images/thumb/d/d8/Phtimecourse.png/569px-Phtimecourse.png]
Measurements are taken every 20 min. These values are the mean of 3 readings. The corresponding error bars represent an interval of twice the standard deviation across the 3 data points centred around the mean value.
| Data | Notes | Date Uploaded |
|---|---|---|
| [http://partsregistry.org/wiki/images/6/64/BBa_K325219pheffect.xls Media:BBa_K325219pheffect.xls] | Raw data from experiment | 21/10/2010 |
Protocol
- The protocol can be found as Experiment 110.
Coloured outputs
In this section we compare the intensity of the different coloured mutants we developed during our project.
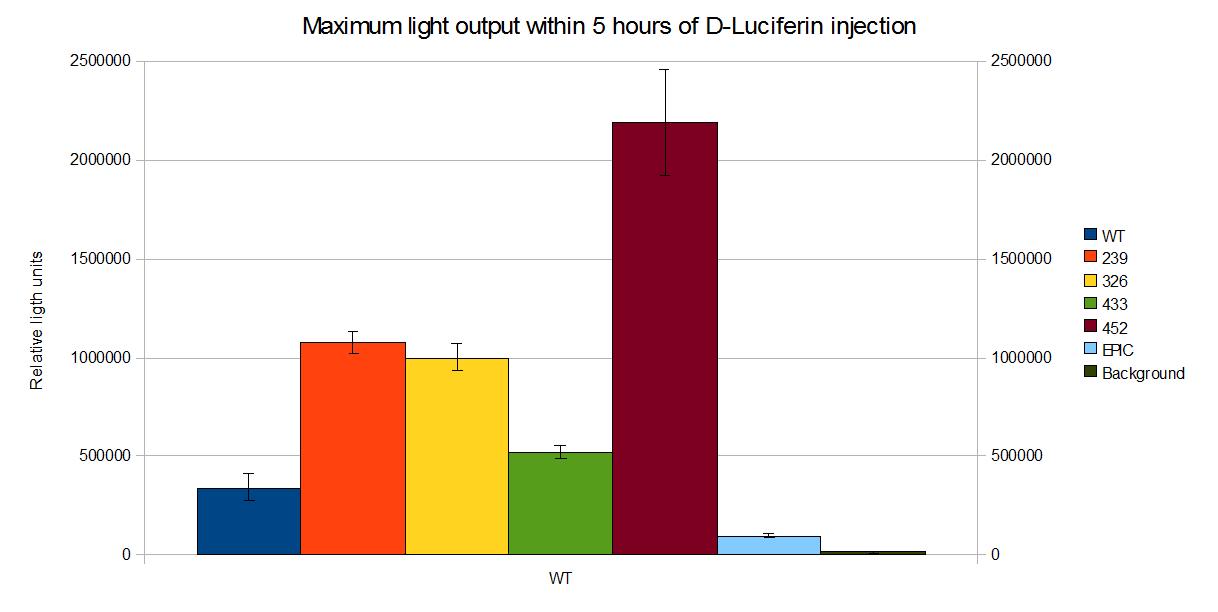
| Data | Notes | Date Uploaded |
|---|---|---|
| CambridgeiGEMcolouredoutputs.xls | Raw data from experiment | 21/10/2010 |
</center>
Compatibility
[http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=cell Chassis:] Device has been shown to work in Top 10 (Invitrogen)
Plasmids: Device has been shown to work on <partinfo>pSB1C3</partinfo>
References
[http://www.ncbi.nlm.nih.gov/pubmed/18949818 [1]:] S.M. Marques and J.C.G. Esteves da Silva, (2009) Firefly Bioluminescence: A Mechanistic Approach of Luciferase Catalyzed Reactions,Life 61, 6-17.
[http://www.nature.com/nature/journal/v440/n7082/abs/nature04542.html [2]:] T. Nakatsu et al. (2006) Structural Basis for the spectral difference in luciferase bioluminescence, Nature 440(16), 372-376.
[http://www.ncbi.nlm.nih.gov/pubmed/11457857 [3]:] K. Gomi and N. Kajiyama, (2001) Oxyluciferin, a Luminescence Product of Firefly Luciferase, Is Enzymatically Regenerated into Luciferin, The Journal of Biological Chemistry, 276(39), 36508-36513.
 "
"