Team:Cambridge/Bioluminescence/Colour
From 2010.igem.org
EmilyKnott (Talk | contribs) (→Results) |
|||
| Line 3: | Line 3: | ||
The peak emission wavelength of luciola cruciata luciferase can be altered by single amino acid changes as reported by [http://www.ncbi.nlm.nih.gov/pubmed/1946326 Kajiyama and Nakano]. Different coloured outputs, as well as being aesthetically pleasing, allow the most appropriate wavelength of output for a particular detector to be chosen. By putting different coloured luciferases under promoters which respond to different inputs, co-reporter assays would be possible. | The peak emission wavelength of luciola cruciata luciferase can be altered by single amino acid changes as reported by [http://www.ncbi.nlm.nih.gov/pubmed/1946326 Kajiyama and Nakano]. Different coloured outputs, as well as being aesthetically pleasing, allow the most appropriate wavelength of output for a particular detector to be chosen. By putting different coloured luciferases under promoters which respond to different inputs, co-reporter assays would be possible. | ||
| - | |||
==Methods== | ==Methods== | ||
Revision as of 23:03, 27 October 2010

The peak emission wavelength of luciola cruciata luciferase can be altered by single amino acid changes as reported by [http://www.ncbi.nlm.nih.gov/pubmed/1946326 Kajiyama and Nakano]. Different coloured outputs, as well as being aesthetically pleasing, allow the most appropriate wavelength of output for a particular detector to be chosen. By putting different coloured luciferases under promoters which respond to different inputs, co-reporter assays would be possible.
Methods
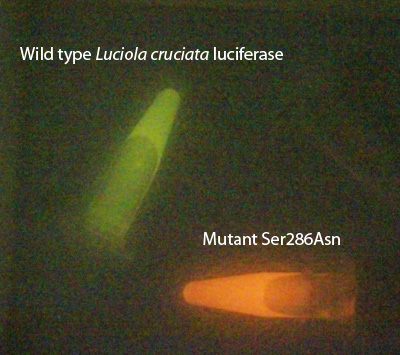
We used site directed mutagenesis to produce these specific mutant luciferase genes. From DNA 2.0 we ordered the sequence of a red-shifted mutant with a Ser286Asn codon change. The luciferase produces an orange glow as shown. The construct was then mutated back to the wild type gene then different point mutations were introduced. pSB1C3 with the luciferase and LRE genes is a sizeable plasmid so we performed two separate PCR reactions to generate the mutagenised construct in two halves (using oligonucleotides with the point mutation). PCR products were run on an agarose gel and the relevant bands gel extracted then joined by Gibson Assembly. Cells were transformed with the product. This method avoids cells being transformed with the non-mutated, template plasmid. As an additional check colonies which grew from successfully transformed cells were imaged inside a dark box after addition of luciferin to check the colour of the light produced was as expected.
Results
We have created biobricks for Luciola cruciata luciferases of five different colours.
| Part | Mutation | Colour | λmax(nm) |
| [http://partsregistry.org/Part:BBa_K325229 BBa_K325229] | Val239Ile | Green | 558 |
| [http://partsregistry.org/Part:BBa_K325209 BBa_K325209] | WT | Yellow/green | 562 |
| [http://partsregistry.org/Part:BBa_K325219 BBa_K325219] | Ser286Asn | Red/Orange | 607 |
| [http://partsregistry.org/Part:BBa_K325239 BBa_K325239] | Gly326Ser | Scarlet | 609 |
| [http://partsregistry.org/Part:BBa_K325249 BBa_K325249] | His433Ser | Red | 612 |
| [http://partsregistry.org/Part:BBa_K325239 BBa_K325259] | Pro452Ser | Yellow | 595 |
The bricks above are the luciferases with the Luciola cruciata luciferin regenerating enzyme under the control of an arabinose inducible promoter. The red/orange Ser286Asn mutant luciferase was also submitted with a promoter but no LRE and without a promoter or the LRE.
 "
"