Team:Heidelberg/Project/Mouse Infection
From 2010.igem.org
(→Introduction) |
|||
| Line 1: | Line 1: | ||
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single}} |
| + | {{:Team:Heidelberg/Single_Pagetop|Project_Mouse_Infection}} | ||
| + | |||
| + | {{:Team:Heidelberg/Side_Top}} | ||
| + | __TOC__ | ||
| + | <br/> | ||
| + | <br/> | ||
| + | === construct schemes === | ||
| + | <br/> | ||
| + | [[Image:PBS SV40 luc2.png|frameless|center|250px|'''Figure 1: Positive control construct.''']] | ||
| + | <br/> | ||
| + | [[Image:PBS SV40 luc2 miR122.png|frameless|center|250px|'''Figure 2: Off-targeting construct.''']] | ||
| + | <br/> | ||
| + | [[Image:PBS_H1_shR_hAAT.png|frameless|center|250px|'''Figure 3: Tuning construct.''']] | ||
| + | <br/> | ||
| + | [[Image:PBS SV40 luc2 hAAT.png|frameless|center|250px|'''Figure 4: Tuned construct.''']] | ||
| + | <br/> | ||
| + | [[Image:PBS TetR miR122 4x.png|frameless|center|250px|'''Figure 5: Repressor construct.''']] | ||
| + | <br/> | ||
| + | [[Image:PBS SV40 TetO2 luc2.png|frameless|center|250px|'''Figure 6: Operator construct.''']] | ||
| + | {{:Team:Heidelberg/Side_Bottom}} | ||
<html> | <html> | ||
<a name="top"></a> | <a name="top"></a> | ||
| Line 10: | Line 30: | ||
</style> | </style> | ||
</html> | </html> | ||
| - | |||
=<i>in vivo</i> study= | =<i>in vivo</i> study= | ||
==Abstract== | ==Abstract== | ||
| Line 46: | Line 65: | ||
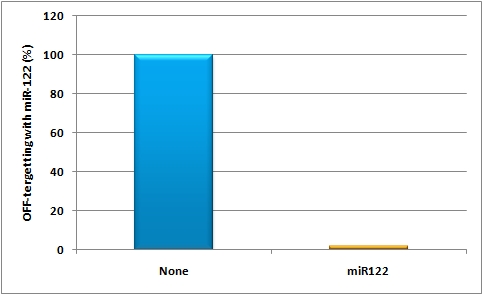
| - | [[Image:Mir122_off_targetting.jpg|thumb|500px|center|miR122 OFF-targetting]] | + | [[Image:Mir122_off_targetting.jpg|thumb|500px|center|miR122 OFF-targetting]]<br/> |
[[Image:HAAT_tuning.jpg|thumb|500px|center|hAAT tuning]] | [[Image:HAAT_tuning.jpg|thumb|500px|center|hAAT tuning]] | ||
| Line 69: | Line 88: | ||
Zincarelli, C., S. Soltys, et al. (2008). "Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection." Mol Ther 16(6): 1073-1080.<br> | Zincarelli, C., S. Soltys, et al. (2008). "Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection." Mol Ther 16(6): 1073-1080.<br> | ||
| - | {{:Team:Heidelberg/ | + | {{:Team:Heidelberg/Single_Bottom}} |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Revision as of 21:22, 27 October 2010

in vivo studyAbstractGene therapy offers a great tool for treatment of various diseases. Nevertheless it is only useful if it allows for specific, fine-tuning of the gene of interest (GOI). IntroductionGene therapy offers a great tool for treatment of various diseases such as failure, muscular dystrophies, and cystic fibrosis (Zincarelli, Soltys et al. 2008). ResultsconstructsThe in vivo analysis should enlighten our gene therapy approach using AAV tropism as well as miRNA binding sites as trigger for expression. The following constructs have been subcloned separately into the AAV context to accomplish those tasks:
mice injectionThe tail vein injection was chosen so as to assess AAV serotype tissue tropism; the luciferase transgene was used for visualizing the relative vector distribution in all the animals in a real-time manner. This allows for in vivo imaging and time lapse experiments. bioluminescence imagingDiscussionMethodscontructsproduction of recombinant virusThe viruses were produced in HEK 293-T cells and purified on an iodixanol gradient according to the virus production protocol. Before infection, the titer of the viruses was quantified using quantitative realtime PCR. procedure involving animalsThe mouse experiments were conducted in accordance with the animal facility of the German Cancer Research Center in Heidelberg. Female NMRI mice were obtained from a collaboration with Dr. Oliver Müller. At 8-10 weeks of age, the animals were injected in the tail vein (TV), with ~ 1x1011 particles of AAV-SV40-luciferase in 200µl of 1x phosphate-buffered saline. The mice are transferred to a holding device which restrains the mouse while allowing access to the tail vein. The tails were warmed before the injections and injections were carried out using 27 gauge needles. All the mice recoverd from the injection quickly without loss of mobility or interruption of grooming activity [http://2010.igem.org/Team:Heidelberg/Project/Mouse_Infection#References (Zincarelli et al., 2008)]. in vivo animal imagingMice were anesthesized in an isofluran chamber. The mice were injected intraperitoneally with 200µl of a 30 mg/ml concentration of D-luciferin. This injection starts the luminescence of luc2. Mice were measured for one to seven minutes post injection under the in vivo bioluminometer. ReferencesGrimm, D. (2002). "Production methods for gene transfer vectors based on adeno-associated virus serotypes." Methods 28(2): 146-157.
|
|||
 "
"