Team:TU Delft/Project/sensing/results
From 2010.igem.org
Lbergwerff (Talk | contribs) (→Does pCaiF really work under starvation conditions???) |
Lbergwerff (Talk | contribs) (→BBa_K398326 pCaiF IN NUMBERS) |
||
| Line 42: | Line 42: | ||
According to our results, there is indeed a difference between both conditions. This is especially clear when the initial glucose concentration is 2 g/L. For the Laurate growth phase in the diauxic experiment we saw a remaining activity from the glucose starvation period and during Laurate consumption we saw a reduction in the GFP production as we expected. | According to our results, there is indeed a difference between both conditions. This is especially clear when the initial glucose concentration is 2 g/L. For the Laurate growth phase in the diauxic experiment we saw a remaining activity from the glucose starvation period and during Laurate consumption we saw a reduction in the GFP production as we expected. | ||
| - | =====[http://partsregistry.org/Part:BBa_K398326 BBa_K398326 pCaiF] | + | =====[http://partsregistry.org/Part:BBa_K398326 BBa_K398326 pCaiF] in numbers===== |
In order to convert Fluorescence arbitrary units to something meaningful, we used the parameters suggest in the part registry for [http://partsregistry.org/Part:BBa_E0040 E0040], we divided the fluorescence random units by 79.429 (conversion factor to nM), then we calculated back the number of moles per well knowing that our reaction volume was 100 uL. If we multiply the total amount of moles per well by the [http://en.wikipedia.org/wiki/Avogadro_constant Avogadro's constant], we will get the total amount of GFP molecules produced in each data point. Then we applied a linear regression to each growth phase and we divided by the O.D. at 600nm in order to normaize the result ([http://partsregistry.org/RiPS RiPS]/O.D.). | In order to convert Fluorescence arbitrary units to something meaningful, we used the parameters suggest in the part registry for [http://partsregistry.org/Part:BBa_E0040 E0040], we divided the fluorescence random units by 79.429 (conversion factor to nM), then we calculated back the number of moles per well knowing that our reaction volume was 100 uL. If we multiply the total amount of moles per well by the [http://en.wikipedia.org/wiki/Avogadro_constant Avogadro's constant], we will get the total amount of GFP molecules produced in each data point. Then we applied a linear regression to each growth phase and we divided by the O.D. at 600nm in order to normaize the result ([http://partsregistry.org/RiPS RiPS]/O.D.). | ||
Revision as of 20:03, 27 October 2010

Sensing Results
pCaiF strength
[http://partsregistry.org/Part:BBa_K398326 BBa_K398326 pCaiF]
[http://partsregistry.org/Part:BBa_K398326 BBa_K398326 (pCaiF)] is by far the smallest part of our project; as we explained before we looked for a promoter that could enable the expression of proteins under low glucose concentrations in order to mimic a diauxic shift for the alkane degradation system: once glucose becomes a limiting factor, the expression of alkane degradation genes under pCaiF control will (theoreticallly) enable the cells to shift from glucose metabolism to alkane degradation. This all could be achieved just by a adding a piece of DNA of just 51 base pairs.
The pCaiF regulation mechanism is really simple, [http://partsregistry.org/Part:BBa_K398326 BBa_K398326 (pCaiF)] contains a cAMP-crp binding domain, cAMP-crp is known as transcriptional regulator. When glucose concentrations are high, cAMP levels are low because there is a lot of energy source that can be metabolized by the cells; however during starvation periods cAMP levels increase and this also increases the concentration of the complex cAMP-crp. This activates at least 180 genes related to starvation response, among which a protein used during Carnitine anaerobic metabolism.
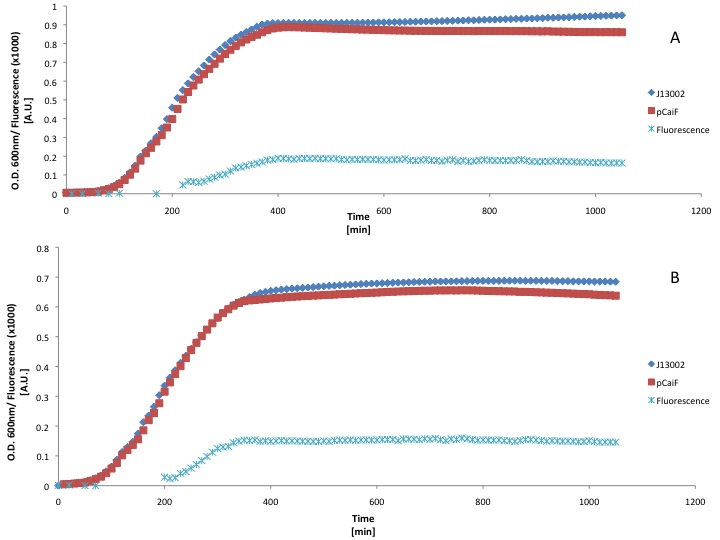
LB medium
Due to time constraints we couldn't make a more elegant circuit or express our proteins using [http://partsregistry.org/Part:BBa_K398326 BBa_K398326 (pCaiF)], however we wanted to show that the part does what we expected: Protein production at low glucose concentrations. We attached a GFP generator in order to measure the output of our part. First we used a rich medium (LB) and we diluted it with M9 medium without glucose (50% v/v) in order to show the differences in protein expression when the carbon source concentration is lower in a rich medium.
Our findings suggest that there is a slight improvement on GFP production when the LB medium is diluted 50%. However the fluorescence produced is a really low signal and we didn't see an increase of GFP production during the stationary phase (starvation). This could be due to the fact that LB medium can keep the cAMP levels low for long periods of time.
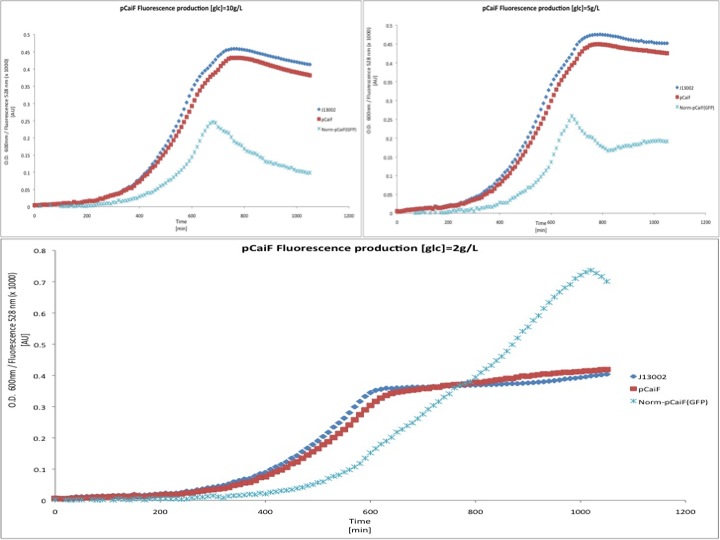
M9 minimal medium
We also tested the response of pCaiF using minimal M9 medium at glucose concentrations of 10 g/L, 5 g/L and 2 g/L. From this experiment we expected to see how pCaiF responds when the starvation phase starts earlier than normal during E. coli cultures.
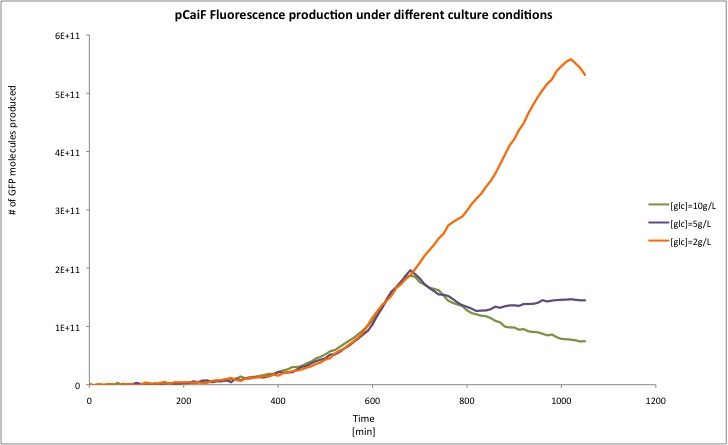
In this experiment we clearly saw a significant increase in GFP production when the glucose initial concentration was 2 g/L compared to our result for an initial glucose concentration of 10 g/L; whereas at 5 g/L we found a slight increase in GFP production at the end of our measurements. From these plots we can conclude that our part is sensitive to cAMP levels as we expected. Moreover, we found that our part is active under starvation periods (stationary phase at low glucose concentrations). We overlapped the plots in order to show the differences between the different conditions tested.
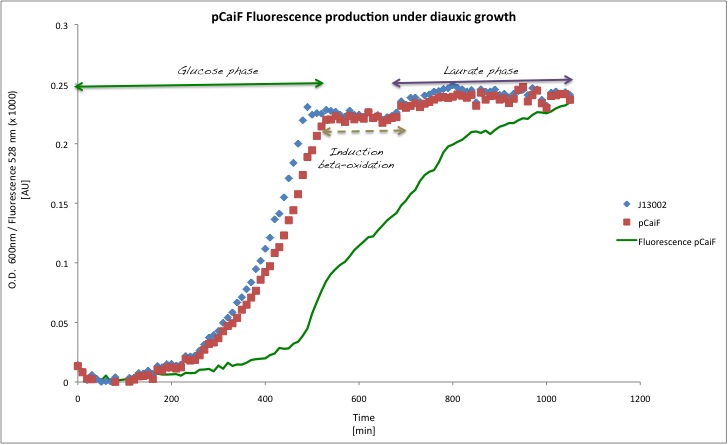
Diauxic shift
In order to see the effect of a secondary carbon source in the medium, we decided to lower the initial glucose amount to 1 g/L and we added Potassium Laurate at a final concentration of 5 mM. With this experiment we expected to see how the GFP profile behaves when cAMP levels drop in presence of a second carbon source.
Our findings in this experiment were really interesting, because as we expected the GFP production decreases when the catabolism of the secondary carbon source starts. You can check that in the plot because of the change in the slope for the GFP profile.
Does [http://partsregistry.org/Part:BBa_K398326 pCaiF] really work under starvation conditions???
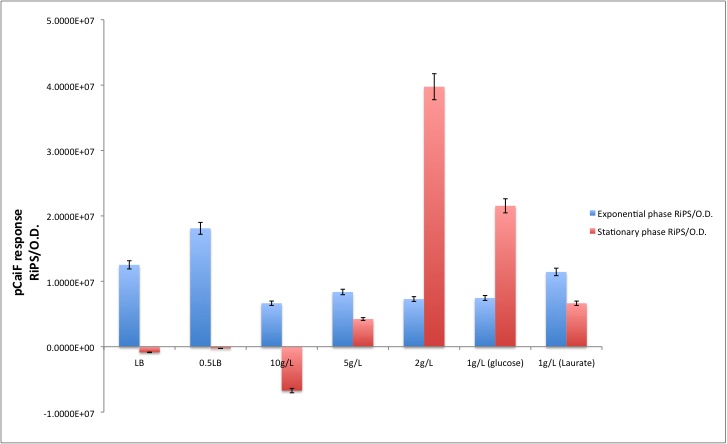
We had some doubts about the activity of pCaiF under starvation periods because we saw a leaky production of GFP that followed the biomass profile. We decided to compare the GFP production during the exponential phase and the stationary phase.
According to our results, there is indeed a difference between both conditions. This is especially clear when the initial glucose concentration is 2 g/L. For the Laurate growth phase in the diauxic experiment we saw a remaining activity from the glucose starvation period and during Laurate consumption we saw a reduction in the GFP production as we expected.
[http://partsregistry.org/Part:BBa_K398326 BBa_K398326 pCaiF] in numbers
In order to convert Fluorescence arbitrary units to something meaningful, we used the parameters suggest in the part registry for [http://partsregistry.org/Part:BBa_E0040 E0040], we divided the fluorescence random units by 79.429 (conversion factor to nM), then we calculated back the number of moles per well knowing that our reaction volume was 100 uL. If we multiply the total amount of moles per well by the [http://en.wikipedia.org/wiki/Avogadro_constant Avogadro's constant], we will get the total amount of GFP molecules produced in each data point. Then we applied a linear regression to each growth phase and we divided by the O.D. at 600nm in order to normaize the result ([http://partsregistry.org/RiPS RiPS]/O.D.).
| Condition | Exponential phase [GFP molecules/O.D.] | Stationary phase [GFP molecules/O.D.] |
|---|---|---|
| LB | 1.2516E+07 | -8.6245E+05 |
| 0.5LB | 1.8101E+07 | -2.5945E+05 |
| [glc]=10g/L | 6.6456E+06 | -6.7109E+06 |
| [glc]=5g/L | 8.3673E+06 | 4.2339E+06 |
| [glc]=2g/L | 7.2869E+06 | 3.9755E+07 |
| Diauxic growth (glucose phase) | 7.4517E+06 | 2.1543E+07 |
| Diauxic growth (Laurate phase) | 1.1435E+07 | 6.6428E+06 |
NOTE: Check at the part registry the definition of [http://partsregistry.org/RiPS RiPS]
Properly, we should express everything in terms of [http://partsregistry.org/PoPS Polymerase Per Second] (PoPS); however we lack of data about the output of B0032 and a standard promoter growing the cells in M9 medium. Thus, the mRNA production by this promoter remains unknown; nevertheless with this results we are reporting the protein production using a Promoter-RBS combination that could become standard for other teams that would decide to express their proteins under glucose starvation.
We developed a simple model, in order to explain in silico what happens with the cAMP levels. If you want to know more about this check our in silico work. ;-)
We reported here the biological function of a new promoter for the part registry that works under starvation periods regulated by cAMP levels, a very useful part for future teams. However, some data is still missing like the mRNA production by this part. Other teams interested on this part should measure this parameter in order to make a complete characterization of our part.

 "
"