Team:Stockholm/20 October 2010
From 2010.igem.org
m |
|||
| Line 131: | Line 131: | ||
**Wash two times with DNA wash buffer | **Wash two times with DNA wash buffer | ||
**Eluate two times in 70µl dH<sub>2</sub>O | **Eluate two times in 70µl dH<sub>2</sub>O | ||
| + | |||
| + | =Johan= | ||
| + | |||
| + | ==Protein A overexpression== | ||
| + | |||
| + | The OD of two overnight cultures of protein A (different colonies) was checked, they had OD 2,8 and 2,6! | ||
| + | |||
| + | I first diluted them 4 times, but then decided that there should be too many dead cells and I diluted it further 750 µl in 50 ml LB. | ||
| + | |||
| + | I then overexpressed the cells with IPTG and then run an tris-gel, Nina has more details. | ||
{{Stockholm/Footer}} | {{Stockholm/Footer}} | ||
Latest revision as of 18:51, 27 October 2010
Contents |
Andreas
IgG protease assay
Me and Elisabeth designed an assay for investigating the IgG protase activity of our BioBrick. In short, the idea behind the assay is as follows:
- Protein extract is prepared from IPTG-induced cells overexpressing IdeS (IgG protease).
- α-mouse IgG-peroxidase goat IgG secondary antibodies (Sigma-Aldrich) are bound to mouse IgG-Agarose (Sigma-Aldrich) beads.
- Excess/unbound secondary antibody are removed by washing with PBS.
- Protein extract is added and left to incubate ON.
- This step allows for digestion of IgG-peroxidase, thereby releasing the peroxidase from the agarose beads.
- After spinning down agarose beads, supernatant is collected.
- Peroxidase substrate is added to identify presence of released peroxidase in the supernatant.
Procedures
See protocols page.
Nina
Protein A overexpression
I induced with IPTG an overnight cuture of 12 ml Protein A.His (N terminal) inserted in the overexpression vector (pEX).
- Samples were taken at 0, 1, 2 & 3 h.
- Pelleted by 1 min of centrifugation at 13 000 rpm.
- Resuspened with 50 ul water and added additional 50 ul of RDSB (Sample buffer with DTT). All samples were stored in the freezer until the induction of 3 hours was done.
- Sonicated for 40 seconds.
- Heated at 95 °C for 5 min.
- Centrifuged 30 seconds at 13 000 rpm.
- Supernatants were added in a gel.
Protein A on Tris-gel
I found out that protein A Z domain that I am working with is not possible to observe on an usual polyacrylamide gel since it is very small (7kDa), I would need to run the overexpression samples on a Tris-gel.
The gel I used was a Tris-gel 10-20% from invitrogen.
Arragement on gel:
After the gel was done I left in a box on shake in coomassie blue staining overnight.
Glycerol Stock
I made a glycerol stock of Protein A.His (N terminal) in the pEX vector.
- 400 ul gycerol
- 800 ul overnight sample
Mimmi
SOD activity
- Start culture from ON culture
- 40ml LBamp
- 400µl old culture
- pEX.SOD
- pEX.yCCS
- At OD=6.0, add IPTG 1mM
- take 10ml samples at 0h, 1h and 2h
- Spinn down and remove LB
- Resuspend in 1ml phosphate buffer, pH=7.0
- keep on ice!
- Transfer to eppendorf tube
- Sonicate
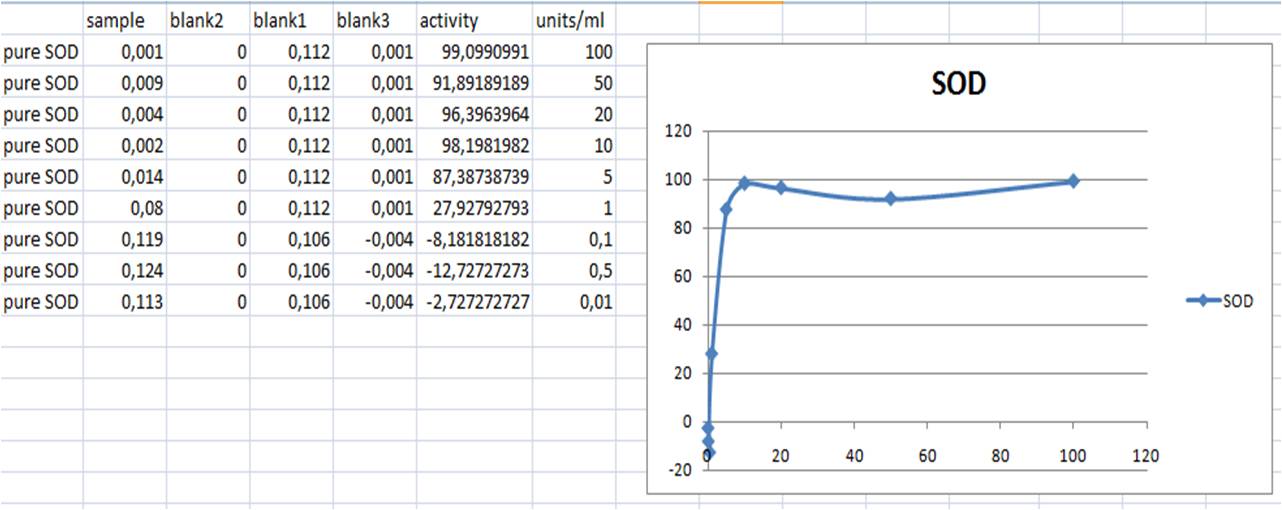
SOD activity standard curve
| mix | samples | blank 1 | blank 3 |
|---|---|---|---|
| sample solution | 20 | ||
| ddH2O | 20 | 20 | |
| WST solution | 200 | 200 | 200 |
| Enzyme solution | 20 | 20 | |
| dilution buffer | 20 | ||
| tot | 240µl | 240µl | 240µl |
- Incubate in 37°C for 20 min
- Measure A440 with nanodrop
- SOD activity (inhibition %) = ((Ablank1 - Ablank3) - (Asample - Ablank2))/(Ablank1 - Ablank3) x 100
plasmid prep
- Follow E.T.Z.N.A plasmid mini prep protocol
- Wash two times with DNA wash buffer
- Eluate two times in 70µl dH2O
Johan
Protein A overexpression
The OD of two overnight cultures of protein A (different colonies) was checked, they had OD 2,8 and 2,6!
I first diluted them 4 times, but then decided that there should be too many dead cells and I diluted it further 750 µl in 50 ml LB.
I then overexpressed the cells with IPTG and then run an tris-gel, Nina has more details.
 |

|
 |

|
 |

|

|

|
 "
"