Team:NYMU-Taipei/Experiments/Speedy switch
From 2010.igem.org
(→Reporting Assay1) |
|||
| Line 19: | Line 19: | ||
=Reporting Assay= | =Reporting Assay= | ||
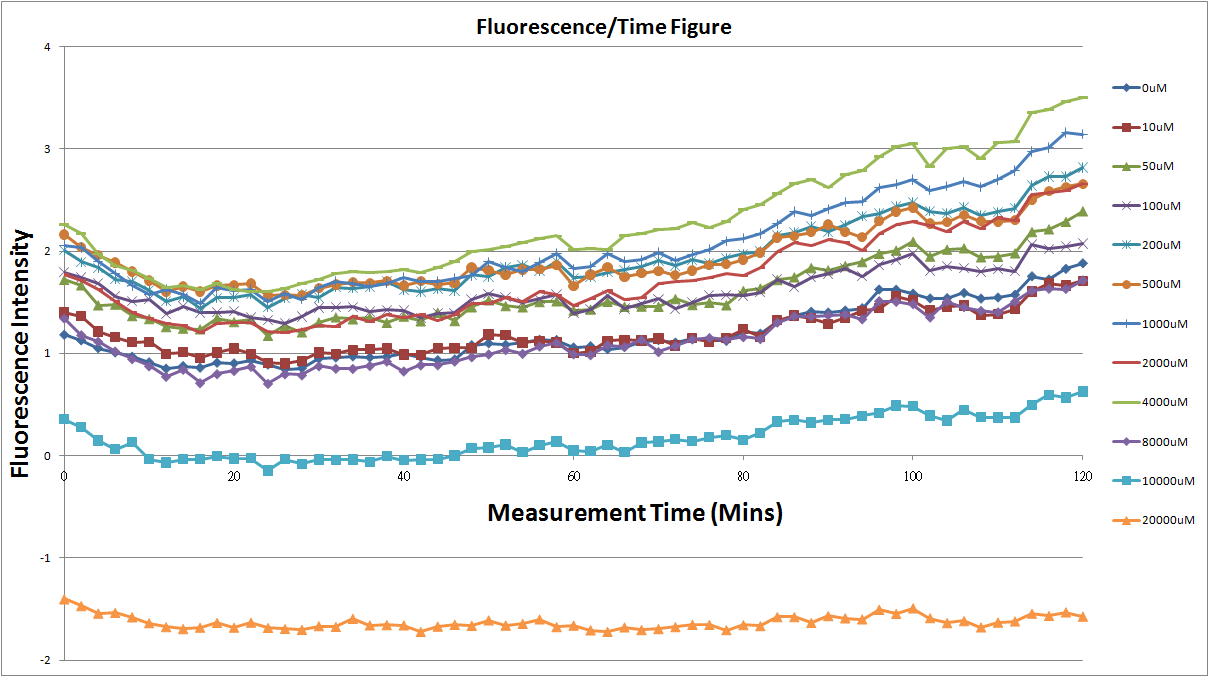
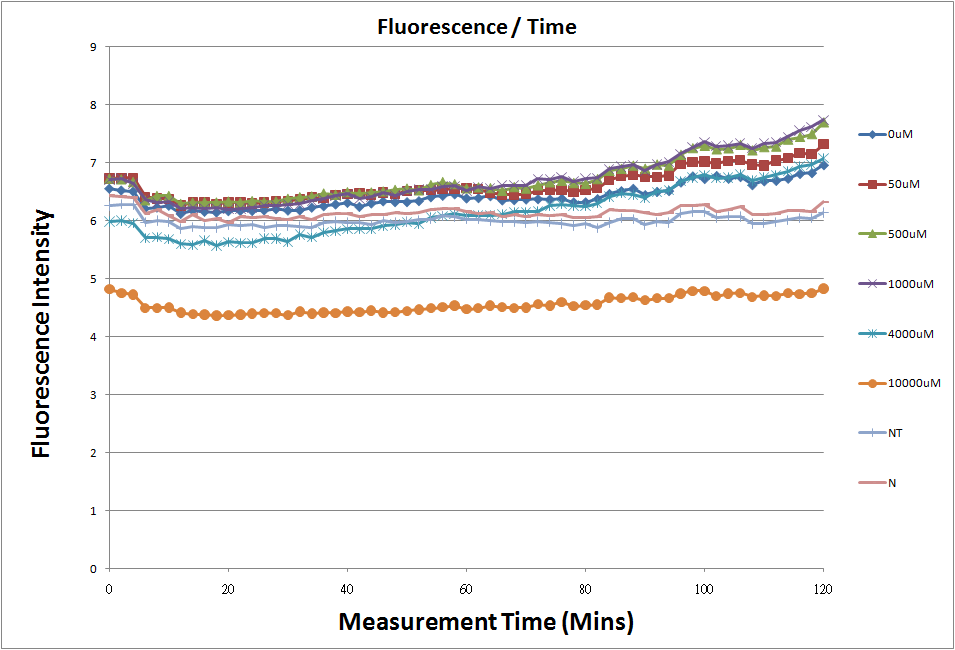
| - | + | Fig.1,fig.3,fig.5,and fig.7 are the original charts of the experiment.N line and NT line are control.N line stands for the sequence didn't have promoter.NT line stands for the sequence didn't have promoter but added 0.1μM theophylline.N line is compared with 0μM and it shows that even though the sequence didn't have promoter it still have little fluorescence so we use it to modify the instrument errors. 0μM line which sequences have promoter but didn’t added theophylline and it still have little fluorescence because mRNA have leaky.and we use this line to find out what’s the degree of mRNA leaky.NT line shows that although it didn’t have promoter it still have little fluorescence.We added theophylline by a solute DMSO and we use this line to modify the other lines which have added different concentration of theophylline. | |
==Reporting Assay1== | ==Reporting Assay1== | ||
*[[https://2010.igem.org/Team:NYMU-Taipei/Experiments/Riboswitch#2010.10.22 reagent formula]] | *[[https://2010.igem.org/Team:NYMU-Taipei/Experiments/Riboswitch#2010.10.22 reagent formula]] | ||
Revision as of 16:23, 27 October 2010
| Home | Project Overview | Speedy reporter | Speedy switch | Speedy protein degrader | Experiments and Parts | Applications | F.A.Q | About Us |
Contents |
Method
- Protocol:
1.Selected genes to be reported are incubated overnight in an LB liquid culture at 37oC and 180-200rpm. This makes sure they are fresh in the morning. Positive and negative controls are also needed.
2.Overnight liquid culture is diluted to OD600 of 0.1, Theophylline is added at concentrations ranging from 0.01mM to 20mM, and the mix incubated for 2-2.5 hours.
3.Measurement of OD at 2 hours: For each used well in the 96-well plate: Take 200uL from the liquid (make sure you pipette this step well) and put it in a cuvette to read the OD600. Note down the OD600 ["OD at 2 hours"], then take the liquid in the cuvette and put it in the right place in the 96-well plate.
4.Measurement of fluorescence: Continuous measurement of fluorescence with the excitation/emission wavelengths 488/511nm for 2 hours, with one fluorescence data point every 2 minutes.
5.Measurement of OD at 4 hours: For each used well in the 96-well plate: Take the liquid from the well and put it in the cuvette to measure the OD ["OD at 4 hours"].
Reporting Assay
Fig.1,fig.3,fig.5,and fig.7 are the original charts of the experiment.N line and NT line are control.N line stands for the sequence didn't have promoter.NT line stands for the sequence didn't have promoter but added 0.1μM theophylline.N line is compared with 0μM and it shows that even though the sequence didn't have promoter it still have little fluorescence so we use it to modify the instrument errors. 0μM line which sequences have promoter but didn’t added theophylline and it still have little fluorescence because mRNA have leaky.and we use this line to find out what’s the degree of mRNA leaky.NT line shows that although it didn’t have promoter it still have little fluorescence.We added theophylline by a solute DMSO and we use this line to modify the other lines which have added different concentration of theophylline.
Reporting Assay1
- The oringinal fluorescence data.
- Normalized the 12 different Theophylline concentration samples with NT & N.
Reporting Assay2
- The original fluorescence data.
- Normalized 12 different concentration of Theophylline with NT & N.
Reporting Assay3
- The oringinal fluorescence data.
- Normalized the 6 different Theophylline concentration samples with NT & N.
Repoting Assay4
- [Reagent Formula]
- The original fluorescence data.
- Normalized 6 different concentration of Theophylline with NT & N.
 "
"