Team:UT-Tokyo/Sudoku modeling
From 2010.igem.org
(→Equations) |
(→Modelling) |
||
| Line 23: | Line 23: | ||
r1,r2,r3,r4 : concentration of 4 recombinass mRNA | r1,r2,r3,r4 : concentration of 4 recombinass mRNA | ||
| + | |||
| + | u1,u2,u3,u4 : concentration of input mRNA | ||
pc,rc : concentration of cre recombinase protein and mRNA | pc,rc : concentration of cre recombinase protein and mRNA | ||
Revision as of 15:34, 27 October 2010


Sudoku
Introduction System Modeling Lab note Result Reference
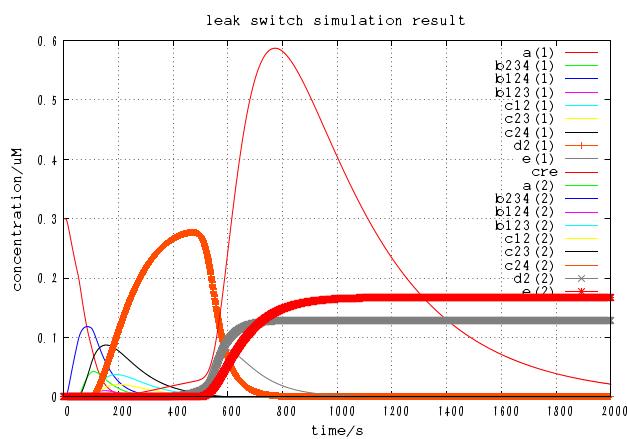
4C3leak switch
We modeled our 4C3 leak switch as ODE system.
Modelling
Variable
Basically, "pn" denotes for protein concentration and "rn" denotes for mRNA concentration
p1,p2,p3,p4 : concentration of 4 recombinase protein
r1,r2,r3,r4 : concentration of 4 recombinass mRNA
u1,u2,u3,u4 : concentration of input mRNA
pc,rc : concentration of cre recombinase protein and mRNA
ps,rs : concentration of SP6 polymerase protein and mRNA
Besides, we treated DNA as continuous variabl, since there could be hundreds of plasmid in one E.coli (multicopy plasmid). There are 32 kinds of variables that stand for DNA corresponding to their different internal states.
Parameters
k0 : mRNA translation rate
k1 : protein degradation rate
k2 : mRNA translation rate
k3 : cre protein degradation rate
l0 : transcription speed per unit concentration of RNA polymearase
l1 : mRNA degradation rate
l2 : cre mRNA degradation rate
K : recombinase binding constant
v : recombinase reaction rate
pT7 : concentration o T7 polymerase
p : terminator leak probability
Equations
We assumed that cre recombinase operate as tetramer, and other recombinase as dimer (but this seems not to be relevant).
Methods
We used our original python program (4 th order explicit Runge-Kutta algorithm) to solve thess equations.
Results
Discussion
conclusion
MS2 phage infection
Whole system
 "
"