Team:ETHZ Basel/Modeling/Combined
From 2010.igem.org
(→Combined Model) |
|||
| Line 2: | Line 2: | ||
{{ETHZ_Basel10_Modeling}} | {{ETHZ_Basel10_Modeling}} | ||
| - | = Combined | + | = Combined Models = |
[[Image:ETHZ_Basel_molecular.png|thumb|400px|'''Combined model.''' Coupled individual models for the simulation of the entire process and their interfaces. The concentration of CheYp determines the movement bias.]] | [[Image:ETHZ_Basel_molecular.png|thumb|400px|'''Combined model.''' Coupled individual models for the simulation of the entire process and their interfaces. The concentration of CheYp determines the movement bias.]] | ||
| - | == | + | == PhyB/PIF3 Light switch - Chemotaxis - Movement == |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
[[Image:ETHZ_Basel_models_overview_comb.png|thumb|400px|'''Combined model.''' Coupled individual models for the simulation of the entire process and their interfaces.]] | [[Image:ETHZ_Basel_models_overview_comb.png|thumb|400px|'''Combined model.''' Coupled individual models for the simulation of the entire process and their interfaces.]] | ||
| - | |||
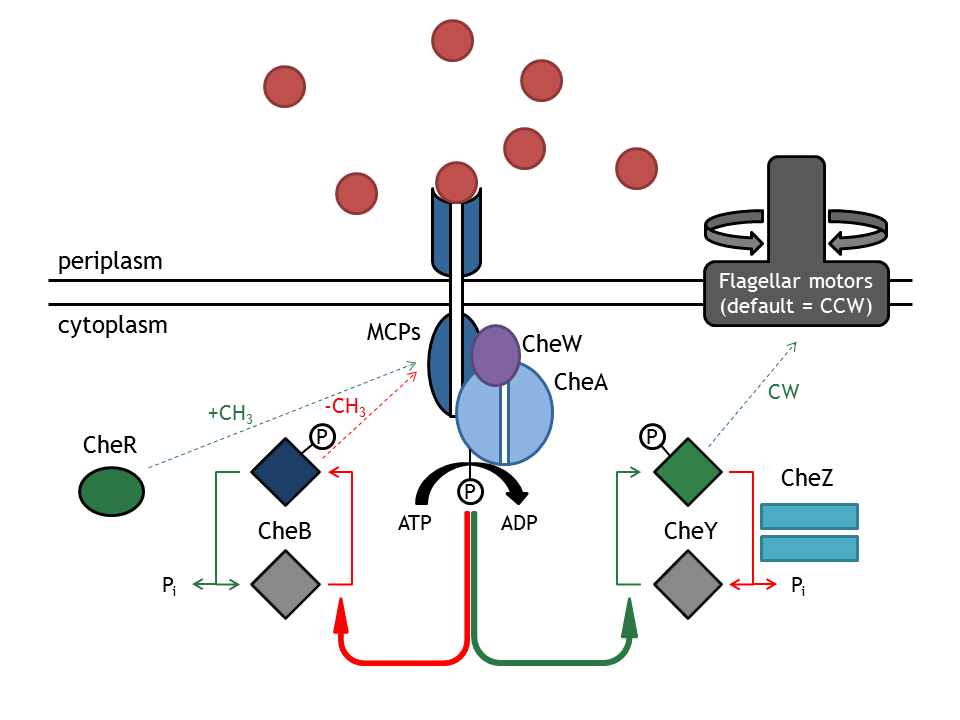
[[Image:ETHZ_Basel_chemotactical_network.png|thumb|400px|'''Schematical overview on the chemotaxis pathway.''' MCPs refers to the membrane receptor proteins and Che are the intracellular proteins of the chemotaxis pathway.]] | [[Image:ETHZ_Basel_chemotactical_network.png|thumb|400px|'''Schematical overview on the chemotaxis pathway.''' MCPs refers to the membrane receptor proteins and Che are the intracellular proteins of the chemotaxis pathway.]] | ||
| - | + | * First, the deterministic light switch and chemotaxis models were combined by assuming a complete removal of a selected Che protein species by the light switch. The interface was defined as the concentration ([LSP-Che]) of this linked device. | |
| - | * | + | * Second, the stochastic movement model was combined with the light switch - chemotaxis model. The interface was defined as the dependency CheYp - directed movement probability (or the bias). At every time point of the simulation, the next state (directed movement/tumbling) of the bacterium was selected, based on the value of the input bias. |
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
The combination of the deterministic and probabilistic models was (first) the biggest challenge and (afterwards) the biggest accomplishment in combining the models. Besides its theoretical complexity, the complete coupling is also conceptually very important. It closes the loop of our E. lemming modeling and it combines two different types of models: deterministic and stochastic, under the same global approach. | The combination of the deterministic and probabilistic models was (first) the biggest challenge and (afterwards) the biggest accomplishment in combining the models. Besides its theoretical complexity, the complete coupling is also conceptually very important. It closes the loop of our E. lemming modeling and it combines two different types of models: deterministic and stochastic, under the same global approach. | ||
<br> | <br> | ||
| Line 35: | Line 19: | ||
<br> | <br> | ||
The main assumption we used in the complete coupling was that the mean run length is a function of the bias, while mean tumbling length is bias - independent. | The main assumption we used in the complete coupling was that the mean run length is a function of the bias, while mean tumbling length is bias - independent. | ||
| + | |||
| + | == Archeal Light switch - Chemotaxis - Movement == | ||
== Download == | == Download == | ||
The combined model is included within the [[Team:ETHZ_Basel/Achievements/Matlab_Toolbox|Matlab Toolbox]] and can be downloaded there. | The combined model is included within the [[Team:ETHZ_Basel/Achievements/Matlab_Toolbox|Matlab Toolbox]] and can be downloaded there. | ||
Revision as of 13:46, 27 October 2010
Combined Models
PhyB/PIF3 Light switch - Chemotaxis - Movement
- First, the deterministic light switch and chemotaxis models were combined by assuming a complete removal of a selected Che protein species by the light switch. The interface was defined as the concentration ([LSP-Che]) of this linked device.
- Second, the stochastic movement model was combined with the light switch - chemotaxis model. The interface was defined as the dependency CheYp - directed movement probability (or the bias). At every time point of the simulation, the next state (directed movement/tumbling) of the bacterium was selected, based on the value of the input bias.
The combination of the deterministic and probabilistic models was (first) the biggest challenge and (afterwards) the biggest accomplishment in combining the models. Besides its theoretical complexity, the complete coupling is also conceptually very important. It closes the loop of our E. lemming modeling and it combines two different types of models: deterministic and stochastic, under the same global approach.
The two models have different timesteps, the molecular one being much faster than the stochastic one, therefore their combination has been done via Simulink interface. Both models have been independently numerically integrated and the output of the deterministic model was passed as an input to the stochastic one, at every slower time-step.
The main assumption we used in the complete coupling was that the mean run length is a function of the bias, while mean tumbling length is bias - independent.
Archeal Light switch - Chemotaxis - Movement
Download
The combined model is included within the Matlab Toolbox and can be downloaded there.
 "
"