Team:Toronto/Protocols
From 2010.igem.org
Disable1987 (Talk | contribs) |
|||
| Line 24: | Line 24: | ||
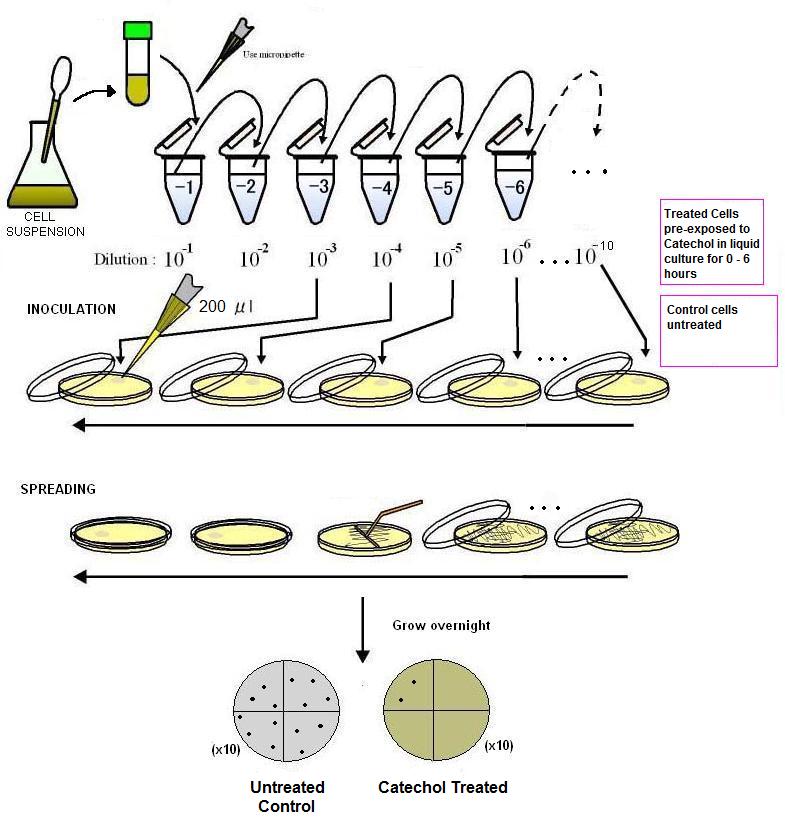
A 0.5 M (10x) stock solution of catechol was made in a 5 ml volume of autoclaved, distilled water. This solution was added to 25 ml of sterile MM2 media to make up a volume of 30 ml. This was added to 20 ml of MM2 innoculated with E.coli DH5a (approximate density 1x10e8 cells per ml) for a final volume of 50 ml (catechol concentration 50mM). Control samples were created as above minus catechol. Treated and untreated samples were placed in a 37C shaker incubator and aliquots were removed every two hours. 0.5 ml aliquots were serially diluted in 4.5 ml of MM2 media and four dilutions representing 1 x 10<sup>-3</sup>, 1 x 10<sup>-4</sup>, 1 x 10<sup>-5</sup> and 1x10<sup>-6</sup> were plated by spreading 200 ul of the diluted sample on LB agar plates. After drying, plates were incubated in an inverted position in a 37C incubator overnight. Plates with between 30 and 300 colonies were counted to determine the number of colony forming units using the method described by Imperial College (iGEM 2009 OpenWetWare). | A 0.5 M (10x) stock solution of catechol was made in a 5 ml volume of autoclaved, distilled water. This solution was added to 25 ml of sterile MM2 media to make up a volume of 30 ml. This was added to 20 ml of MM2 innoculated with E.coli DH5a (approximate density 1x10e8 cells per ml) for a final volume of 50 ml (catechol concentration 50mM). Control samples were created as above minus catechol. Treated and untreated samples were placed in a 37C shaker incubator and aliquots were removed every two hours. 0.5 ml aliquots were serially diluted in 4.5 ml of MM2 media and four dilutions representing 1 x 10<sup>-3</sup>, 1 x 10<sup>-4</sup>, 1 x 10<sup>-5</sup> and 1x10<sup>-6</sup> were plated by spreading 200 ul of the diluted sample on LB agar plates. After drying, plates were incubated in an inverted position in a 37C incubator overnight. Plates with between 30 and 300 colonies were counted to determine the number of colony forming units using the method described by Imperial College (iGEM 2009 OpenWetWare). | ||
| + | |||
| + | [[image:Toronto_Dilutions.jpg|center]] | ||
<h2>'''Making Chemically Competent Cells'''</h2> | <h2>'''Making Chemically Competent Cells'''</h2> | ||
Revision as of 06:29, 27 October 2010
| Home | Project | Design | Protocols | Notebook | Results | Parts Submitted to the Registry | Modeling | Software Used | Human Practices | Safety | Team | Official Team Profile | FAQ | Acknowledgments |
|---|
Contents |
Protocols
Tolerance Assay
A 0.5 M (10x) stock solution of catechol was made in a 5 ml volume of autoclaved, distilled water. This solution was added to 25 ml of sterile MM2 media to make up a volume of 30 ml. This was added to 20 ml of MM2 innoculated with E.coli DH5a (approximate density 1x10e8 cells per ml) for a final volume of 50 ml (catechol concentration 50mM). Control samples were created as above minus catechol. Treated and untreated samples were placed in a 37C shaker incubator and aliquots were removed every two hours. 0.5 ml aliquots were serially diluted in 4.5 ml of MM2 media and four dilutions representing 1 x 10-3, 1 x 10-4, 1 x 10-5 and 1x10-6 were plated by spreading 200 ul of the diluted sample on LB agar plates. After drying, plates were incubated in an inverted position in a 37C incubator overnight. Plates with between 30 and 300 colonies were counted to determine the number of colony forming units using the method described by Imperial College (iGEM 2009 OpenWetWare).
Making Chemically Competent Cells
Day 1
1. Start a 5 mL LB overnight culture in the 37C incubator from either a colony streaked plate or with one of the last frozen competent stocks of the strains you want to make competent.
Day 2
1. Inoculate the 5 mL overnight culture into a new flask with 200 mL of fresh LB.
2. Grow until O.D. 600 = 0.4.
a. To check O.D., take 1 mL of the bacterial culture and put it in a plastic cuvette.
b. Take another plastic cuvette and add in 1 mL of LB.
c. Change the wavelength of the spectrophotometer to 600 nm.
d. Blank using the LB cuvette.
e. Measure and record the absorbance of the cuvette containing the culture.
f. If the O.D. is below 0.4, place the flask back in the 37°C incubator and repeat ‘e’ at a later time point. Dispose of the cuvette with the culture but keep the cuvette with the LB as a blank.
3. Transfer 50 mL of the culture into a pre-chilled Falcon tube. Repeat for each of the remaining 3 Falcon tubes.
4. Place the Falcon tubes in ice for 15 minutes to cool the cultures.
5. Pellet the cells using a Beckman centrifuge at 4000 rpm for 10 minutes at 4°C.
6. Pour out the media in the sink and stand the Falcon tubes in an inverted position on paper towels for 1 min to drain away traces of liquid.
7. Re-suspend the pellets in each tube with 10 mL of ice cold 0.1M CaCl2 by lightly pipetteing with a P1000.
8. Store on ice for 10 min.
9. Pellet the cells using a Beckman centrifuge at 4000 rpm for 10 minutes at 4°C.
10. Pour out the media in the sink and stand the Falcon tubes in an inverted position on paper towels for 1 min to drain away traces of liquid.
11. Re-suspend the pellets in each tube with 2 mL of ice cold 0.1M CaCl2 by lightly pipetteing with a P1000.
12. Pool the aliquots and keep on ice for 1 hour.
13. Make a dry ice/ethanol bath.
a. Dry ice is on the 14th floor in the corresponding room where the incubators are at on our floor.
b. Ethanol is under the fumehood in a cabinet labelled “Flammables” beside the imaging room.
14. Add in 4.8 mL of ice cold 40% glycerol and very gently invert the tube to mix well.
15. Divide the mixture into the pre-chilled microfuge tubes in 400 µL aliquots.
16. Flash freeze the aliquots in the dry ice bath.
17. Store in the -80°C freezer.
Transforming Chemically Competent Cells
1. Thaw a stock of competent cells that you want to transform into on ice for 10 minutes. DO NOT REMOVE IT FROM THE ICE DURING THIS TIME.
2. If you are doing more than 1 transformation, take 100 µL of cells from the freezer stock and add it to prechilled microfuge tubes. If not, take 100 µL of cells to use as transformation controls.
3. Add 1 µL of DNA to the cells you want transformed. Stir gently with the pipette as any serious agitation will decrease transformation by a lot.
4. Leave mixtures on ice for 30 minutes.
5. Heat shock cells for 30s at 42°C. Different times and temperatures will decrease transformation efficiency.
6. Place mixtures on ice for 5 min.
7. Add LB to each tube until the total volume is 1 mL. (i.e. If there is 100 µL of cells, add 900 µL of fresh LB)
8. Place the tubes in the 37°C shaker for 1 hour.
9. Pre-warm and label (date, initials, what DNA is transformed with what cells, amount plated) selection plates in the 37°C incubator with the sign “BACTERIA ONLY”.
10. Spread plate 2 amounts of transformants (10 µL and 50 µL) onto the corresponding selection plates.
11. Place overnight in the 37°C incubator with the sign “BACTERIA ONLY”.
Digesting DNA
1. Thaw the NEBuffer(s) that you will need to digest your DNA of choice.
a. The NEBuffer will have to be chosen carefully if digesting with 2 or more enzymes.
b. Always try to maximize the percent activity (indicated in the NEB info slips for each restriction enzyme).
c. Be on the lookout for star activity in different buffers.
d. If you can’t find a common NEBuffer, you can also digest sequentially.
2. Add your components in the following order (for a 25 µL reaction)
a. ddH2O
b. 10X NEBuffer (2.5 µL)
c. DNA
d. Restriction Enzyme (5 units)
i. If you scale up the reaction volumes, you won’t need to add more enzyme because 5 units is already way more than enough (unless you digesting 5+ µg)
3. Turn on the PCR machine and place the tubes inside one of the 48 well blocks. Select the program Kris > DIGEST, and the block your sample is in (A or B).
a. If you need to run the samples on a gel, add 5 µL of 6x Loading dye and either freeze the sample in -20°C.
4. If the enzymes can be heat inactivated (check the NEB slips), select the program in Kris > INACTIV.
a. If not, use the PCR purification kit to remove the restriction enzyme from the mixture.
b. Inactivation of enzymes is important if you want to ligate your DNA (otherwise, the enzyme will just digest right after the ligase works on it).
Ligating DNA
1. Thaw the 10X T4 DNA Ligase Buffer.
2. Add the components in the following order (10 µL reaction):
a. 10X T4 DNA Ligase Buffer (1 µL)
b. Vector DNA (10 ng)
c. Insert DNA (6 insert:1 vector molar ratio)
d. ddH2O (to make the volume 9.5 µL total, if necessary)
e. T4 DNA Ligase Buffer (0.5 µL)
3. Leave at room temperature for 30 minutes.
4. Freeze the mixture in the -20°C freezer for future use or use 1 µL in a transformation immediately.
Gel Electrophoresis
1. Based on what percentage gel you want to run, add half the percentage in grams to 50 mL of 1X TBE in a 125 mL / 250 mL flask.
a. For example, if you want to make a 1% gel, add in 0.5g of agarose into 50 mL of 1X TBE.
2. Place the flask in the microwave and heat it for 45 seconds. If it starts bubbling, immediately open the microwave door.
3. Take out the flask and swirl the mixture for 5-10 seconds. Use gloves or a folded dry paper towel to hold the flask to avoid getting burned.
4. Place the flask in the microwave and heat for 15-30 seconds. If it starts bubbling, immediately open the microwave door. Let the flask and mixture cool until it is warm when you touch it.
5. Add 2.5 µL of ethidium bromide into the cooling mixture and mix well.
a. Be VERY careful not to get any EtBr on your skin/clothes.
6. Take the small tray and tape both ends. Be sure to wrap the tape tightly around edges and corners so that no fluid will leak out.
7. Slowly pour the gel into the tray and see if it leaks. If it does, try to add more tape in the area to stop the leak.
8. Once the gel is poured, place the comb into preset ridges on the top of the tray.
9. Once the gel has solidified, carefully remove the comb and the tape.
10. Place the gel tray (with the gel still on it) into the electrophoresis cell. Place the wells facing the negative electrode (black).
11. Fill the electrophoresis cell with enough 1X TBE to cover the gel.
12. Add in 10 µL of to the side where the positive electrode (red) is located.
13. Load your samples and ladder(s); record what samples are in what wells.
14. Close the cover of the electrophoresis cell and plug it into the power supply box.
15. Set the voltage to 110V (you can push it to 120V, but don’t go any higher as the gel will melt) and turn off the power supply when the dye reaches at least ¾ down the gel (usually 45 min – 1 hr).
Polymerase Chain Reaction
1. Let the dNTPs, primers, DNA template(s) and 10X reaction buffer thaw at room temperature
2. Keep the Taq polymerase on ice at all times.
3. Add the reagents in the following order for a 25 µL reaction:
a. 10X reaction buffer (2.5 µL)
b. ddH2O (filling volume up to 25 µL, following these values it would be 17.15 µL)
c. Forward primer (0.2 µL)
d. Reverse primer (0.2 uL)
e. 2 mM dNTPs (3.75 µL)
f. DNA template (1 µL)
g. Taq polymerase (0.2 µL)
4. Repeat step 3 for all DNA samples you want amplified. Make the appropriate controls.
a. For the negative control, replace the DNA template with 1 µL of ddH2O.
5. Place the tubes in the PCR machine (in the same block) and select the PCR program you want to run. If you don’t have a pre-made program, make the following program in your own folder:
a. Set the lid temperature to 105°C
b. Step 1: 95°C for 15 min
c. Step 2: 94°C for 30 seconds
d. Step 3: (Primer Tm – 3) for 30 seconds
e. Step 4: 72°C for (1 min per kb of product)
f. Step 5: Repeat steps 2-4 39 times.
g. Step 6: 72°C for 20 min
h. Step 7: 4°C forever
6. Run it in a gel or use the PCR purification kit as necessary.
 "
"