Team:UCSF/Project/Signaling
From 2010.igem.org
| Line 52: | Line 52: | ||
| - | [[Image:GPCR_graph.png| | + | [[Image:GPCR_graph.png|620px]] |
| Line 77: | Line 77: | ||
| - | [[Image:Stronger_signaling_approach2.jpg| | + | [[Image:Stronger_signaling_approach2.jpg|620px]] |
'''''Figure 2''''' | '''''Figure 2''''' | ||
Revision as of 04:38, 27 October 2010
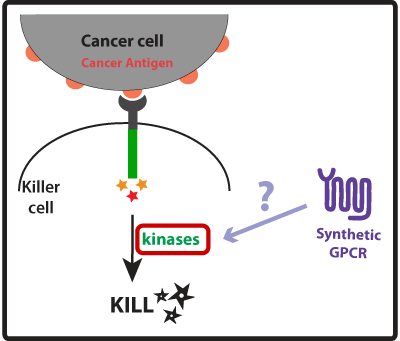
STRONGER SIGNALINGGoal: Engineer a stronger signaling response on killer cells that will lead to a more effective/faster elimination of the cancer cell.
1. Co-stimulation by transfection of killer cells with a synthetic GPCR 2. Reengineer killer cell’s receptors to have multiple activation domains 3. Bypassing steps in the signaling cascade by engineering direct connections between the killer cell’s receptor and downstream effectors.
1. Co-stimulation – we successfully used a device from last year’s UCSF team (registry part# BBA_K209400) and showed enhanced signaling activation 2. Multiple activation domains – we designed and constructed new devices for this approach but did not get to test them 3. Bypassing steps in activation cascade – we designed a few devices for this approach but did not had enough time to assemble them from parts to devices.
In this part of our project we wanted to create devices that made our synthetic cancer killers more effective than their natural counterparts at eliminating cancer cells by enhancing the activation mechanisms that lead to the killing response. Once our synthetic cancer killers confirmed they were in the presence of a cancer cell (using the devices developed in the GREATER PRECISION part of our project) they would take no time to eliminate it and could rapidly move on to the next target. Natural immune killers use receptor-associated intracellular activation domains to tell the inside of the cell that it needs to get ready to kill the cancer cell. This means that these intracellular activation domains, such as ITAMs (Immunoreceptor Tyrosine-based Activation Motifs), once activated recruit and activate kinases and a cascade of signaling events is initiated culminating in the formation of the immune synapse between the killer cell and the cancer cell. The killer cell then releases killing agents into the immune synapse that tell the cancer cell to apoptose (die!). We thought of three ways to improve the activation of the killer cell (described in more detail below): (1) Co-stimulation using a synthetic GPCR to enhance strength of killing response; (2) Engineer receptors with multiple activation domains to increase the level of activation; and (3) Bypass steps on the activation cascade to possibly improve speed of response.
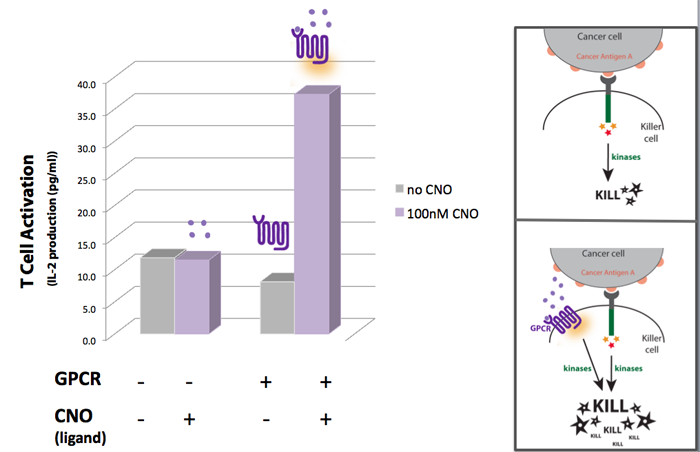
Concept, Experimental design and Results One way we thought of enhancing the killing response was to increase the amount of kinases activated, as these seem to have a central role in the induction of killing on the killer cells. For that we looked into other receptors that once activated would also lead to the activation of kinases. Last year, the UCSF iGEM team used synthetic G protein-coupled receptors (GPCRs) to engineer immune cells to move (chemotax) towards new targets. In the future these synthetic cellbots would be able to detect specific chemicals released by tumors and move towards them more efficiently (for more info on this project please see UCSF iGEM 2009). GPCRs are activated by small molecule ligands instead of cell surface proteins. Because intracellular kinases are involved in chemotaxis, we hypothesized that the synthetic GPCRs used by the 2009 team that showed to mediate chemotaxis could possibly enhance the killing response by increasing the overall amount of kinases activated (Figure 1). Figure 1
A synthetic GPCR shown to mediate immune cell migration towards the ligand CNO (results of UCSF iGEM team 2009 project) was introduced to T cells as a potential way to increase the level of kinase activation. IL-2 production was used as a measure of T-cell activation. The results show that when in the presence of CNO, T cells transfected with the synthetic GPCR , have a higher level of activation than non-transfected T-cells.
Our experimental model suggests a synthetic biology approach to eliminate cancer cells more effectively. If we could engineer synthetic GPCRs that recognize small molecules secreted by cancer cells, we could make killer cells move faster to a cancer site (based on UCSF 2009 project results) AND once at the cancer site our synthetic killers would also be equipped to eliminate the cancer more efficiently. The additional kinase activation caused by the synthetic GPCRs would augment the normal activation signal to enhance the killing response of the killer cell.
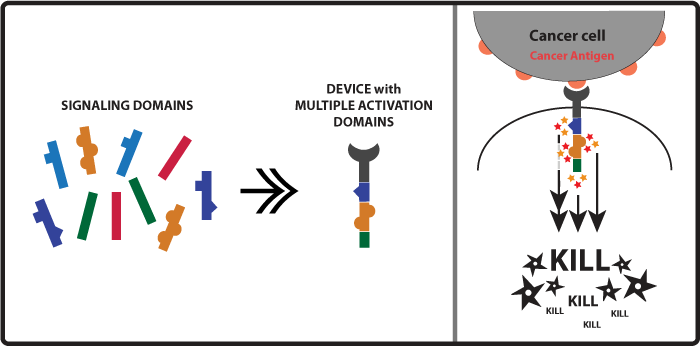
Concept and Experimental design As mentioned in the introduction above when killer cell receptors bind to cancer antigens their intracellular activation domains (such as ITAMs) get activated and lead to the activation of kinases. So another approach to increase the amount of kinases activated and ultimately enhancement of the killing response would be to activate more activation domains. A small input would lead to a amplified output. With this in mind our team came up with the idea of constructing receptors with multiple activation domains so when one receptor would become activated it would lead to the activation of a higher number of kinases (Figure 2). Examples of immune killer cell’s receptors activation domains are CD3zeta, FCRgamma, DAP10. Figure 2
Devices: We were able to build, but not test, the following devices: CAR-CD3z-CD3z CAR-DAP10-CD3z CAR-DAP10-DAP10 CAR-FCRg-CD3z CAR-FCRg-DAP10
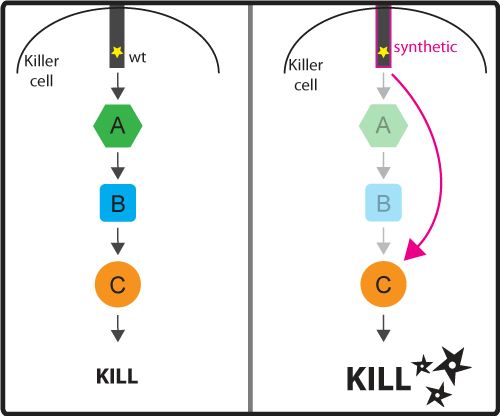
Concept and Design As discussed in the introduction, the activation signaling cascade (or pathway) connect the input (activation by binding cancer antigen) with the ultimate output (cancer cell killing). These signaling cascades while effective at regulating the ultimate response by the killer cell, can be slow because once the ligand binds to the receptor, the signal has to pass through the long chain of molecules that make up the signaling pathway. We would like to engineer a possible faster pathway by bypassing certain intermediate steps of the pathway (for example regulatory steps). As a result, we would cut down on the number of steps needed to be taken before reaching the desired output (Figure 3). Figure 3
|
Right
|
 "
"