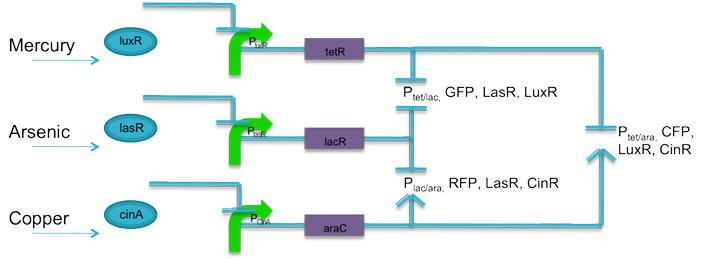
Team:Virginia United/Project
From 2010.igem.org
(Difference between revisions)
(→Optimization of Fluorescent Proteins) |
(→Optimization of Fluorescent Proteins) |
||
| Line 127: | Line 127: | ||
===== Optimization of Fluorescent Proteins ===== | ===== Optimization of Fluorescent Proteins ===== | ||
| - | |||
| - | |||
Optimization of protein expression is an essential skill in synthetic biology; the ability to tweak protein expression to the levels one desires eases the process of designing precise biological systems. Fluorescent proteins function as appropriate applications in optimization experiments because fluorescence expression levels can easily be measured by a spectrofluorometer or plate reader. | Optimization of protein expression is an essential skill in synthetic biology; the ability to tweak protein expression to the levels one desires eases the process of designing precise biological systems. Fluorescent proteins function as appropriate applications in optimization experiments because fluorescence expression levels can easily be measured by a spectrofluorometer or plate reader. | ||
| + | |||
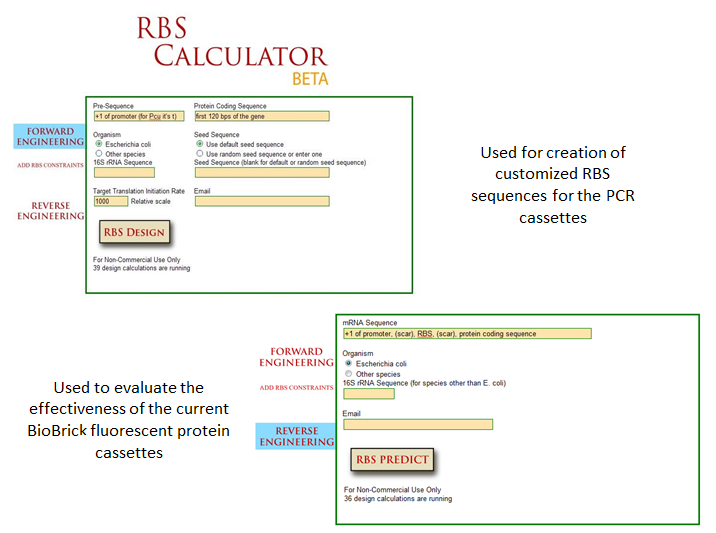
| + | [[Image:RBS Calculator.png|720px|thumb|center]] | ||
One method of optimization that has recently been explored is the inherent differences in efficiency of gene-specific ribosome binding sites (Voigt et al., 2009). The majority of the time when designing a construct in iGEM, a ‘default’ RBS is used by including just the basic Shine-Dalgarno sequence found in all prokaryotic ribosome binding sites. What many do not realize is that when utilizing the Shine-Delgarno conserved sequence in different gene constructs, the efficiency of the expression for that construct depends on the gene. Utilizing the RBS calculator developed by Voigt et al., one can test the actual optimization rates of the ribosome binding sites by measuring fluorescence output of the final constructs. This allows more control in the system, instituting more flexibility in the precise design of one’s constructs. | One method of optimization that has recently been explored is the inherent differences in efficiency of gene-specific ribosome binding sites (Voigt et al., 2009). The majority of the time when designing a construct in iGEM, a ‘default’ RBS is used by including just the basic Shine-Dalgarno sequence found in all prokaryotic ribosome binding sites. What many do not realize is that when utilizing the Shine-Delgarno conserved sequence in different gene constructs, the efficiency of the expression for that construct depends on the gene. Utilizing the RBS calculator developed by Voigt et al., one can test the actual optimization rates of the ribosome binding sites by measuring fluorescence output of the final constructs. This allows more control in the system, instituting more flexibility in the precise design of one’s constructs. | ||
Revision as of 02:21, 27 October 2010
 "
"