Team:ETHZ Basel/Achievements/Technical Standard
From 2010.igem.org
(→Documentation of a new technical standard that supports the design of BioBrick Parts or Devices) |
(→Documentation of a new technical standard that supports the design of BioBrick Parts or Devices) |
||
| Line 4: | Line 4: | ||
= Technical Standard = | = Technical Standard = | ||
== Documentation of a new technical standard that supports the design of BioBrick Parts or Devices== | == Documentation of a new technical standard that supports the design of BioBrick Parts or Devices== | ||
| - | '''Update on BBF | + | '''Update on BBF RFC28 to make it compatible to Tom Knight's original assembly standard''' |
BBF RFC28 is a method for combinatorial multi-part assembly based on the Type II restriction enzyme AarI and it is a highly efficient method for constructing fusion proteins. As we have a great number of constructs to generate, efficiency in the cloning process is vital for the project. Unfortunately it is not compatible with the general iGEM standardized biobricks. As a consequence, the fusion proteins are not compatible with Tom Knight's original assembly standard as they don't contain the required prefix and suffix sequences. | BBF RFC28 is a method for combinatorial multi-part assembly based on the Type II restriction enzyme AarI and it is a highly efficient method for constructing fusion proteins. As we have a great number of constructs to generate, efficiency in the cloning process is vital for the project. Unfortunately it is not compatible with the general iGEM standardized biobricks. As a consequence, the fusion proteins are not compatible with Tom Knight's original assembly standard as they don't contain the required prefix and suffix sequences. | ||
Revision as of 15:52, 26 October 2010
Technical Standard
Documentation of a new technical standard that supports the design of BioBrick Parts or Devices
Update on BBF RFC28 to make it compatible to Tom Knight's original assembly standard
BBF RFC28 is a method for combinatorial multi-part assembly based on the Type II restriction enzyme AarI and it is a highly efficient method for constructing fusion proteins. As we have a great number of constructs to generate, efficiency in the cloning process is vital for the project. Unfortunately it is not compatible with the general iGEM standardized biobricks. As a consequence, the fusion proteins are not compatible with Tom Knight's original assembly standard as they don't contain the required prefix and suffix sequences.
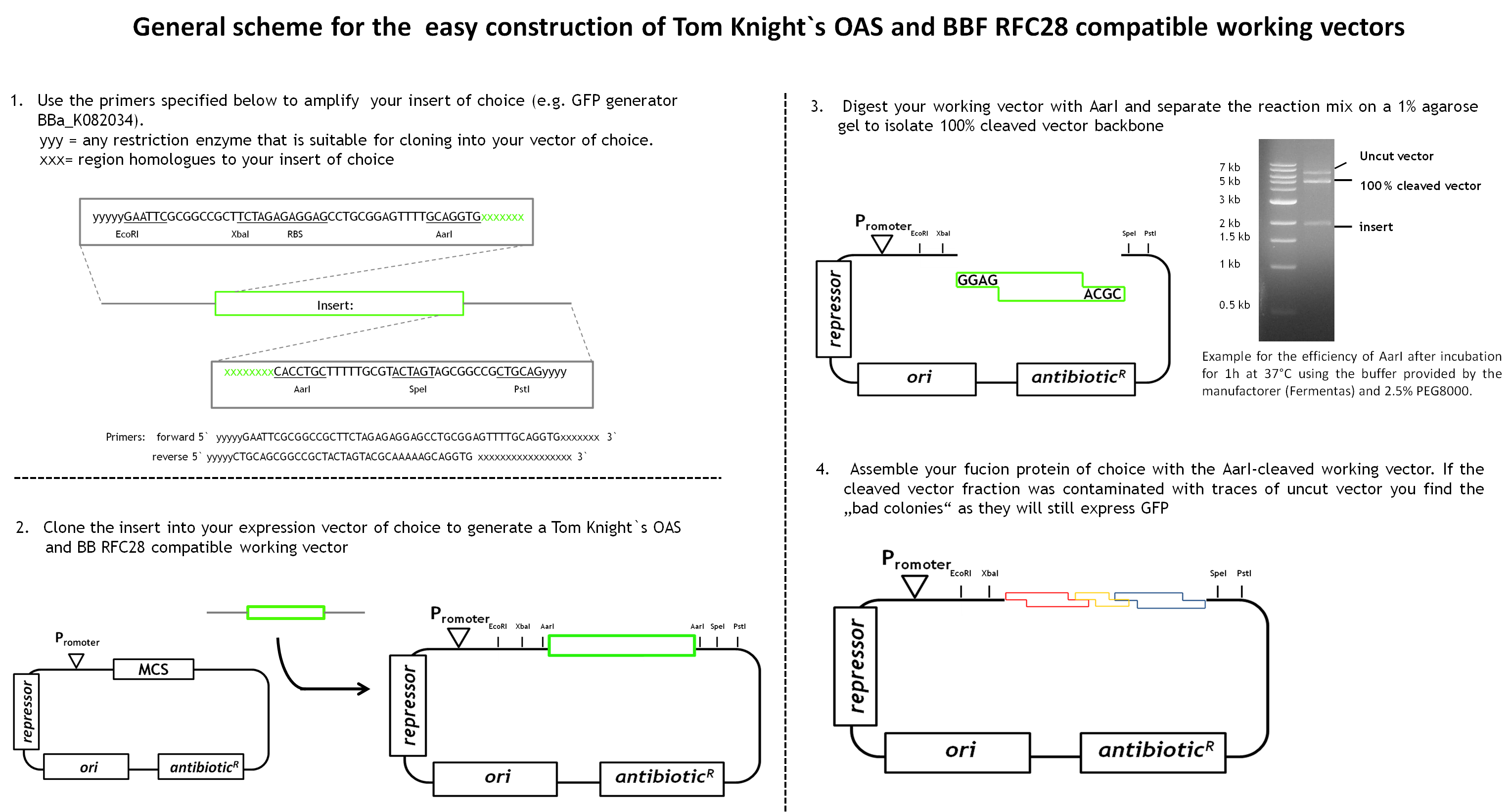
During our project we learned a lot about the cloning of fusion proteins using the BBF RFC28 assembly method and how to make it work efficiently. We would like to share our experience with you, especially as we worked out a general design method for the facilitated construction of Tom Knight's OAS and BBF RFC28 compatible working vectors (the expression vector that is finally used for the assembly) that can be used for the final assembly step. Our vector design includes a GFP-mediated screening system for screening clones containing the fusion protein.
As we had already started the cloning procedure before learning about the little difficulties of the assembly method and before making up our minds on how to improve it and link it to the Tom Knight's BBF, our fusion proteins were not cloned into a Tom Knight's OAS-compatible working vector. Nevertheless, the design scheme that is outlined below is under construction.
In general a working vector that can be used for efficient assembly needs to contain an insert that is flanked by AarI-sites. Those cleavage sites must be designed such that AarI-digestion releases the insert and generates a vector backbone with 5` overhangs compatible with the BBF RFC28 standard. The rational behind cloning an insert into the vector instead of simply adding two AarI sites is that AarI does not work with the same high efficiency on every cleavage site. Therefore, cleaved vector can be efficiently separated from uncut vector fractions by separation on a 1% agarose gel. By choosing a constitutively expressed reporter protein as insert, positive clones containing the assembled protein can be distinguished from clones containing the original insert (traces of uncut vector might still be present in the cleaved vector fraction). Constitutive expression of the reporter is very convenient as adding an inducer to the selection plates is not necessary. This gives total freedom of choice for the fusion protein expression system. By designing the primers for amplification of the insert such that the insert in the end contains the Tom Knight` OAS suffix and prefix sequences plus a ribosome binding site makes any available expression vector readily usable as standardized assembly vector. the insert simply needs to be cloned into the chosen expression vector. The standard primer sequences for the amplification of the insert and the general scheme for the working vector construction are outlined below.
 "
"