Team:ETHZ Basel/Modeling/Chemotaxis
From 2010.igem.org
(→Modeling of the chemotaxis pathway) |
|||
| Line 20: | Line 20: | ||
For the combined model implementation, only two chemotaxis models have been further investigated to focus on implementation and extension of the original models: Spiro et al. (1997) and Mello & Tu (2003). | For the combined model implementation, only two chemotaxis models have been further investigated to focus on implementation and extension of the original models: Spiro et al. (1997) and Mello & Tu (2003). | ||
| - | == Spiro et al. (1997) [[Team:ETHZ Basel/Modeling/Chemotaxis#References|[1]]] == | + | == Model based on Spiro et al. (1997) [[Team:ETHZ Basel/Modeling/Chemotaxis#References|[1]]] == |
| - | The model | + | The model based on Spiro et al. (1997) [[Team:ETHZ Basel/Modeling/Chemotaxis#References|[1]]] has been used to identify candidates of the chemotaxis receptor pathway by enabling removal or addition of a species upon light induction. For all Che proteins (CheR, Y, B, Z), the concentrations stay below/above the threshold, until they are added again (LSPs are deactivated with far-red light and dimerization is reversed). In terms of aspartate ligand concentration, the best results were obtained for assuming a high ligand concentration (saturation, the methylation level of the receptors is high). For CheY and Z, the reaction times are much faster than for CheB and CheR. |
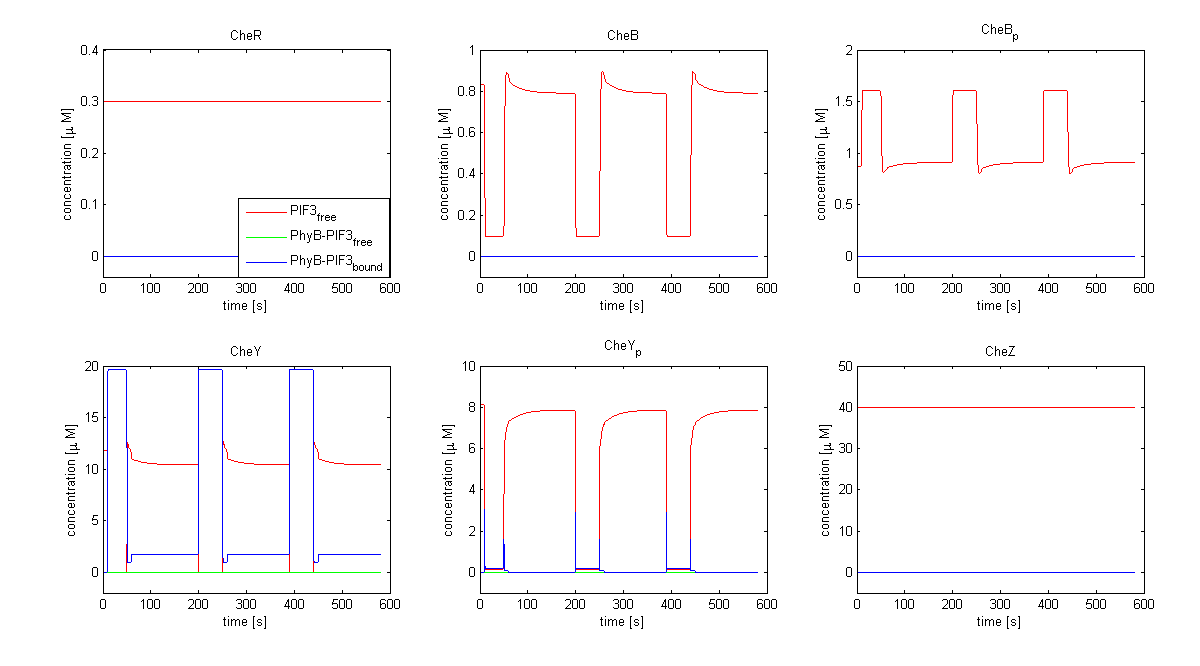
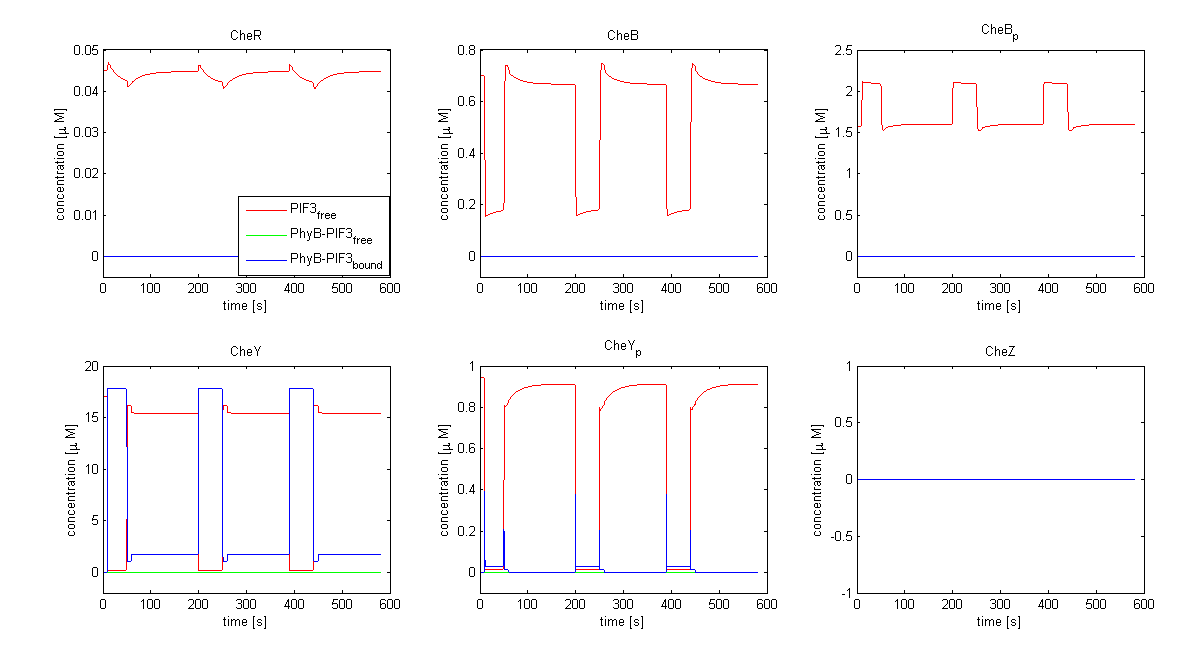
| - | [[Image:ETHZ_Basel_chemotaxis_spiro_1.png|thumb|center|833px|'''Che protein species predicted by the model | + | [[Image:ETHZ_Basel_chemotaxis_spiro_1.png|thumb|center|833px|'''Che protein species predicted by the model based on Spiro et al. (1997)''' CheY is coupled to PIF3. PhyB is present in a concentration of 100 uM, anchor in a concentration of 130 uM. Medium asparate levels (10^-6 uM) were chosen.]] |
{| border="0" align="center" | {| border="0" align="center" | ||
|- valign="top" | |- valign="top" | ||
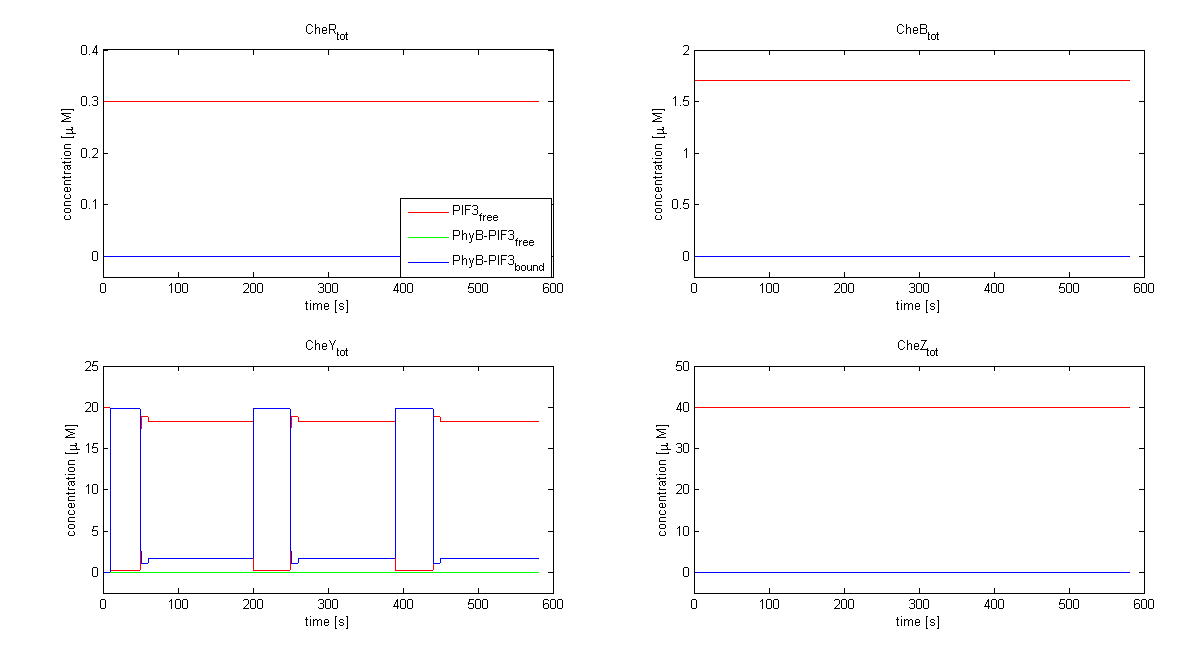
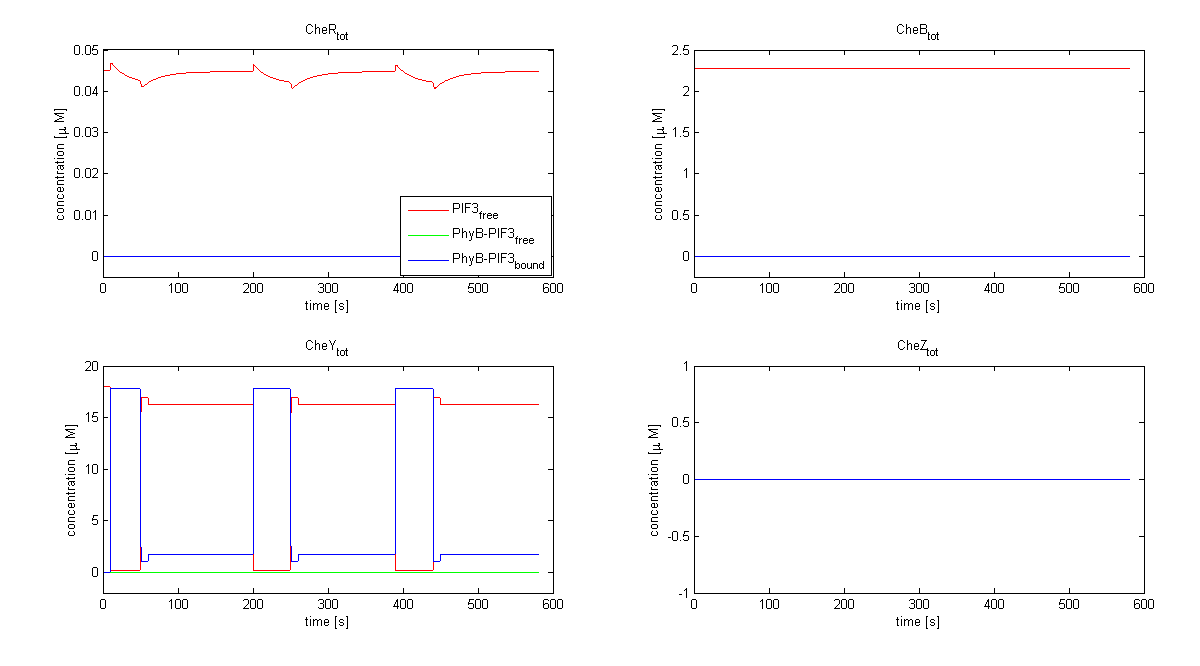
| - | |[[Image:ETHZ_Basel_chemotaxis_spiro_2.png|thumb|center|550px|'''Total Che protein species predicted by the model | + | |[[Image:ETHZ_Basel_chemotaxis_spiro_2.png|thumb|center|550px|'''Total Che protein species predicted by the model based on Spiro et al. (1997)''' Only the total concentration of CheY is changed by the light switch in this model.]] |
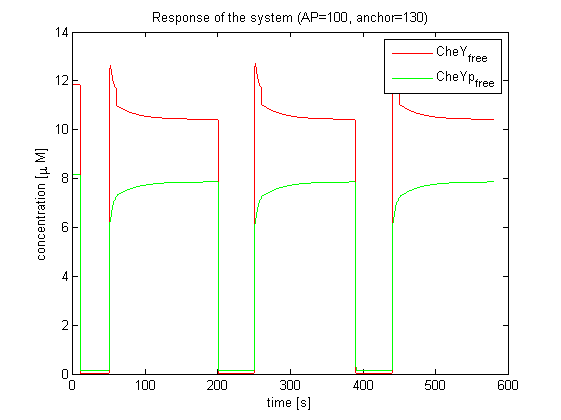
| - | |[[Image:ETHZ_Basel_chemotaxis_spiro_3.png|thumb|250px|'''Response of the system.''' CheYp amplitude is predicted to be high, according to the model | + | |[[Image:ETHZ_Basel_chemotaxis_spiro_3.png|thumb|250px|'''Response of the system.''' CheYp amplitude is predicted to be high, according to the model based on Spiro et al. (1997)]] |
|} | |} | ||
| - | == Mello & Tu (2003) [[Team:ETHZ Basel/Modeling/Chemotaxis#References|[2]]] == | + | == Model based on Mello & Tu (2003) [[Team:ETHZ Basel/Modeling/Chemotaxis#References|[2]]] == |
| - | The model | + | The model based on Mello & Tu (2003) differs from the one based on Spiro et al. (1997) in a way, that it is able to reach perfect and near-perfect adaptation. Mello & Tu investigated robustness of the chemotaxis receptor network, covering the effect of attractant binding through the phosphorylation of CheY. Governing ODEs are derived by applying the law of mass action to the known reactions. Five states of methylation and demethylation of the attractant-bound and free receptors are considered. The model based on Mello & Tu (2003) shows similar behavior compared to the adapted Spiro et al. (1997) model. |
| - | [[Image:ETHZ_Basel_chemotaxis_mello_1.png|thumb|center|833px|'''Che protein species predicted by the model | + | [[Image:ETHZ_Basel_chemotaxis_mello_1.png|thumb|center|833px|'''Che protein species predicted by the model based on Mello & Tu (2003)''' CheY is coupled to PIF3. PhyB is present in a concentration of 100 uM, anchor in a concentration of 130 uM. Medium asparate levels (10^-6 uM) were chosen.]] |
{| border="0" align="center" | {| border="0" align="center" | ||
|- valign="top" | |- valign="top" | ||
| - | |[[Image:ETHZ_Basel_chemotaxis_mello_2.png|thumb|center|550px|'''Total Che protein species predicted by the model | + | |[[Image:ETHZ_Basel_chemotaxis_mello_2.png|thumb|center|550px|'''Total Che protein species predicted by the model based on Mello & Tu (2003).''' The total concentration of CheR is changed in addition to the CheYp concentration in this model]] |
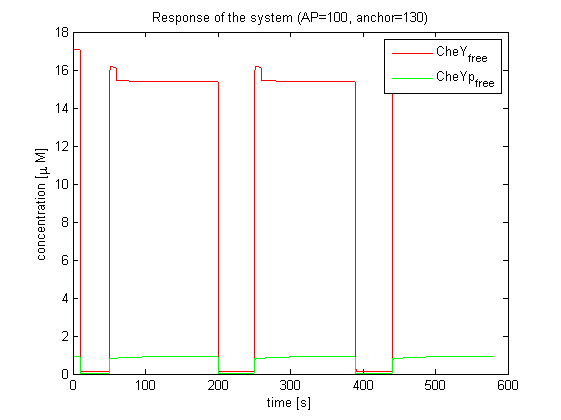
| - | |[[Image:ETHZ_Basel_chemotaxis_mello_3.png|thumb|250px|'''Response of the system.''' The model | + | |[[Image:ETHZ_Basel_chemotaxis_mello_3.png|thumb|250px|'''Response of the system.''' The model based on Mello & Tu (2003) predicts a much smaller CheYp amplitude than the model based on Spiro et al. (1997)]] |
|} | |} | ||
== References == | == References == | ||
Revision as of 11:26, 26 October 2010
Modeling of the chemotaxis pathway
Background
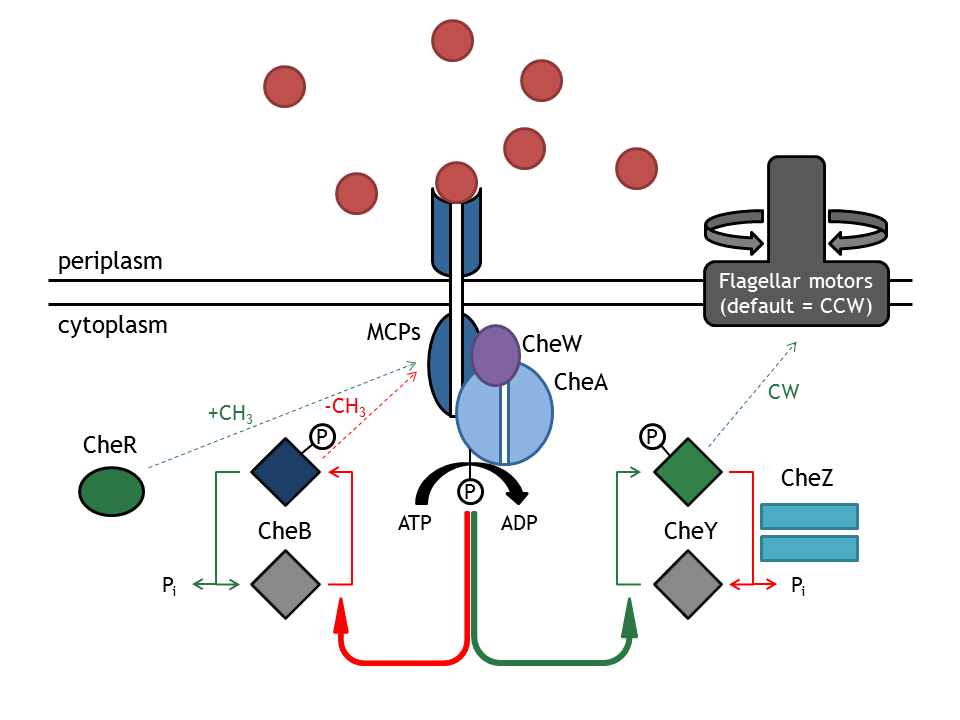
The chemotaxis receptor pathway in E. coli is quite complex. Published models of chemotaxis [1], [2], [3], [4] thus have to use many assumptions in order to answer the investigated question. In the case of E. lemming, the question regarding the chemotaxis pathway is:
- How does the output species (CheYp bias) react to perturbations of upstream species?
The chemotaxis network represents the main decision factor in bacterial movement and therefore, it received special attention for the experimental design: It was decided to adapt and extend four different models based on published approaches [1], [2], [3], [4] to be able to achieve a more general consensus prediction of the chemotaxis behavior in E. lemming.
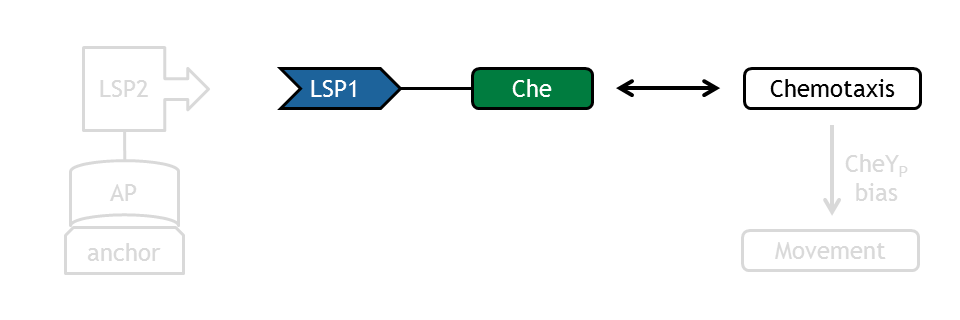
The interface to the light switch model is the selected Che species, which is linked to one light-sensitive protein (LSP1). Concentration of this species (LSP1-Che) is determined by the light switch model. Evaluation of the model based on this approach was conducted by changing concentration of selected Che species according to a series of time steps, reflecting light pulses. This assumes total removal of the species. In addition to the selected Che species (CheR, B, Y, Z), possible phosphorylated subspecies were analyzed.
Important for analysis of the chemotaxis network in E. lemming is the concentration of the output species CheYp. Threshold of suitable CheYp concentration is determined, according to predictions of the movement model, regarding an optimization of corresponding tumbling / directed movement frequency. Response of the chemotaxis models was measured by taking the relative amplitude in CheYp concentration between two different light pulses. For CheR and Y, the difference in concentration of CheYp drops more than Delta (initial value - threshold), for CheB and Z it increases more than Delta. Manipulation of CheR and Y concentration therefore have an inverse effect on tumbling / directed movement ratio than CheB and Z.
For the combined model implementation, only two chemotaxis models have been further investigated to focus on implementation and extension of the original models: Spiro et al. (1997) and Mello & Tu (2003).
Model based on Spiro et al. (1997) [1]
The model based on Spiro et al. (1997) [1] has been used to identify candidates of the chemotaxis receptor pathway by enabling removal or addition of a species upon light induction. For all Che proteins (CheR, Y, B, Z), the concentrations stay below/above the threshold, until they are added again (LSPs are deactivated with far-red light and dimerization is reversed). In terms of aspartate ligand concentration, the best results were obtained for assuming a high ligand concentration (saturation, the methylation level of the receptors is high). For CheY and Z, the reaction times are much faster than for CheB and CheR.
Model based on Mello & Tu (2003) [2]
The model based on Mello & Tu (2003) differs from the one based on Spiro et al. (1997) in a way, that it is able to reach perfect and near-perfect adaptation. Mello & Tu investigated robustness of the chemotaxis receptor network, covering the effect of attractant binding through the phosphorylation of CheY. Governing ODEs are derived by applying the law of mass action to the known reactions. Five states of methylation and demethylation of the attractant-bound and free receptors are considered. The model based on Mello & Tu (2003) shows similar behavior compared to the adapted Spiro et al. (1997) model.
References
[1] [http://www.pnas.org/content/94/14/7263.full Spiro et al: A model of excitation and adaptation in bacterial chemotaxis. PNAS 1997 94;14;7263-7268.]
[2] [http://www.cell.com/biophysj/retrieve/pii/S0006349503700216 Mello & Tu: Perfect and Near-Perfect Adaptation in a Model of Bacterial Chemotaxis. Biophysical Journal 2003 84;5;2943-2956.]
[3] [http://www.plosbiology.org/article/info:doi/10.1371/journal.pbio.0020049 Rao et al: Design and Diversity in Bacterial Chemotaxis. PLoS Biol 2004;2;2;239-252.]
[4] [http://www.nature.com/nature/journal/v387/n6636/abs/387913a0.html Barkai & Leibler: Robustness in simple biochemical networks. Nature 1997;387;913-917.]
 "
"