Team:ETHZ Basel/Achievements
From 2010.igem.org
(Difference between revisions)
(→MATLAB Toolbox (Lemming Toolbox)) |
(→MATLAB Toolbox (Lemming Toolbox)) |
||
| Line 14: | Line 14: | ||
* Image aquisition - to receive images either from a microscope (live image stream) or to work with saved images in a storage device | * Image aquisition - to receive images either from a microscope (live image stream) or to work with saved images in a storage device | ||
* [https://2010.igem.org/Team:ETHZ_Basel/InformationProcessing/CellDetection Cell Detection & Tracking] - to detect cells in the image streams and keep track of their trajectories through out the image stream | * [https://2010.igem.org/Team:ETHZ_Basel/InformationProcessing/CellDetection Cell Detection & Tracking] - to detect cells in the image streams and keep track of their trajectories through out the image stream | ||
| - | * Microscope control - to adjust the position of the stage to capture the cells properly, to shine red/far-red light on the cells to influence their movement | + | * [https://2010.igem.org/Team:ETHZ_Basel/InformationProcessing/Microscope Microscope control] - to adjust the position of the stage to capture the cells properly, to shine red/far-red light on the cells to influence their movement |
* Input devices - to capture inputs from a joystick or a keyboard | * Input devices - to capture inputs from a joystick or a keyboard | ||
| - | * Controller - to select a cell & to drive it to a target | + | * [https://2010.igem.org/Team:ETHZ_Basel/InformationProcessing/Controller Controller] - to select a cell & to drive it to a target |
| - | * Models (for simulations) | + | * [https://2010.igem.org/Team:ETHZ_Basel/Modeling Models] (for simulations) |
| - | ** Molecular Model - capturing & integrating the biological pathways into the simulations | + | ** [https://2010.igem.org/Team:ETHZ_Basel/Modeling/Combined Molecular Model] - capturing & integrating the biological pathways ([https://2010.igem.org/Team:ETHZ_Basel/Modeling/Chemotaxis chemotaxis pathway] & the [https://2010.igem.org/Team:ETHZ_Basel/Modeling/Light_Switch light switch]) into the simulations |
| - | ** Movement Model - simulates the motion of the E .lemming | + | ** [https://2010.igem.org/Team:ETHZ_Basel/Modeling/Movement Movement Model] - simulates the motion of the E .lemming |
** Image generator - to generate a realistic look & feel for the simulations | ** Image generator - to generate a realistic look & feel for the simulations | ||
| - | * Visualization | + | * [https://2010.igem.org/Team:ETHZ_Basel/InformationProcessing/Visualization Visualization] |
** Cell Visualization - outputs the visuals combining the microscope images and the outputs of above Simulink blocks | ** Cell Visualization - outputs the visuals combining the microscope images and the outputs of above Simulink blocks | ||
| - | ** E. lemming 2D - the game | + | ** [https://2010.igem.org/Team:ETHZ_Basel/InformationProcessing/Game E. lemming 2D] - the game |
The complete toolbox can be downloaded here [add link]. Please visit the [https://2010.igem.org/Team:ETHZ_Basel/Achievements/Matlab_Toolbox MATLAB Toolbox page] for further information | The complete toolbox can be downloaded here [add link]. Please visit the [https://2010.igem.org/Team:ETHZ_Basel/Achievements/Matlab_Toolbox MATLAB Toolbox page] for further information | ||
Revision as of 14:19, 25 October 2010
Achievements Overview
BioBrick Toolbox
MATLAB Toolbox (Lemming Toolbox)
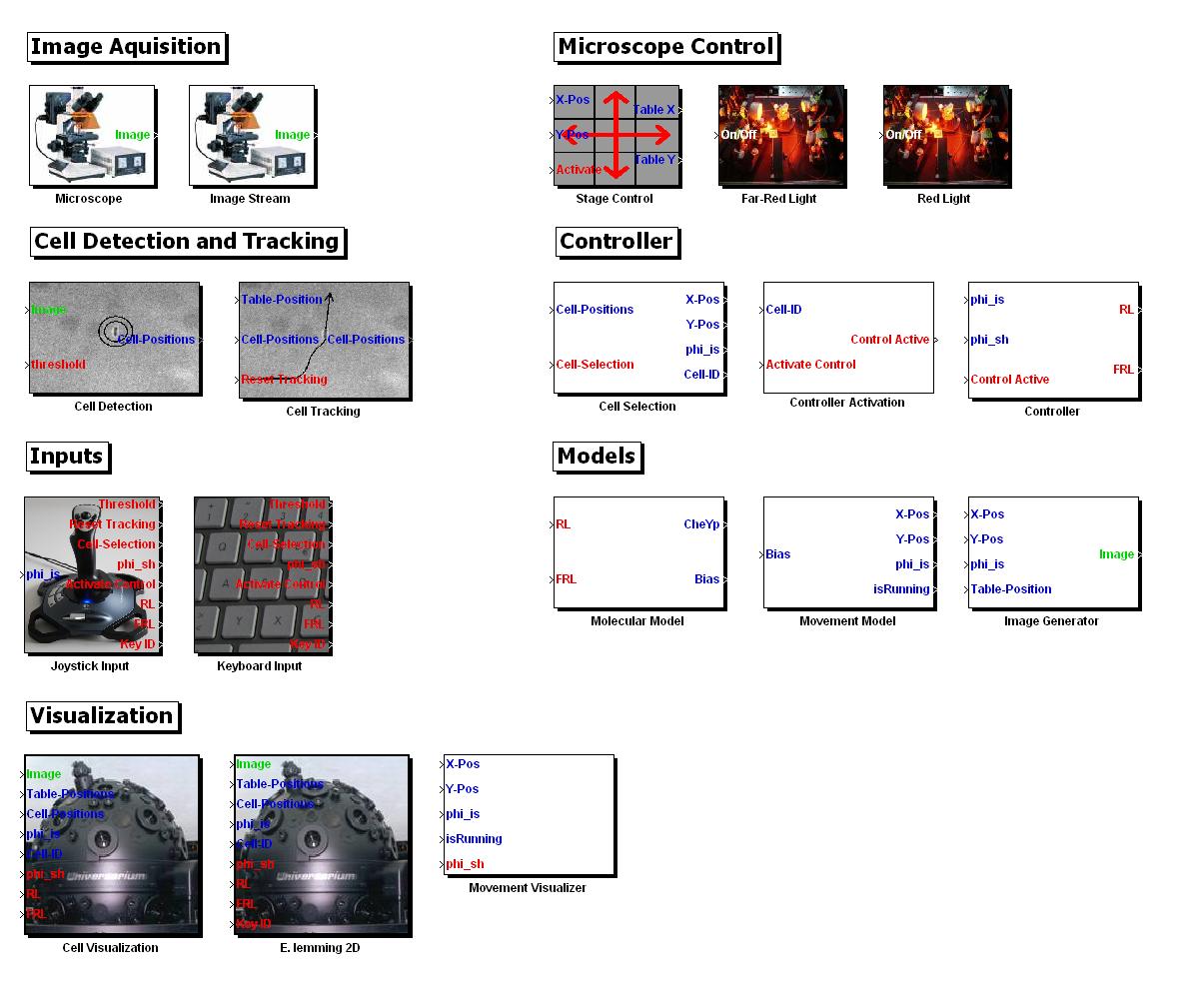
MATLAB Toolbox Screenshot of the Simulink blocks available in the toolbox. Please note that these blocks can be visually assembled with the Simulink blocks of other Toolboxes (e.g. blocks for nearly any mathematical function), and that the algorithms we developed can be accessed directly through Matlab code, too.
As a result of our efforts to make all our computer based tools & algorithms modular and re-usable, we have assembled the entire in-silico setup of the E. Lemming into a MATLAB toolbox. This includes the following Simulink modules that encapsulate the algorithms we used.
- Image aquisition - to receive images either from a microscope (live image stream) or to work with saved images in a storage device
- Cell Detection & Tracking - to detect cells in the image streams and keep track of their trajectories through out the image stream
- Microscope control - to adjust the position of the stage to capture the cells properly, to shine red/far-red light on the cells to influence their movement
- Input devices - to capture inputs from a joystick or a keyboard
- Controller - to select a cell & to drive it to a target
- Models (for simulations)
- Molecular Model - capturing & integrating the biological pathways (chemotaxis pathway & the light switch) into the simulations
- Movement Model - simulates the motion of the E .lemming
- Image generator - to generate a realistic look & feel for the simulations
- Visualization
- Cell Visualization - outputs the visuals combining the microscope images and the outputs of above Simulink blocks
- E. lemming 2D - the game
The complete toolbox can be downloaded here [add link]. Please visit the MATLAB Toolbox page for further information
 "
"


