Team:Stockholm/12 October 2010
From 2010.igem.org
(→Andreas) |
NinaSchiller (Talk | contribs) (→Andreas) |
||
| Line 1: | Line 1: | ||
{{Stockholm/Top2}} | {{Stockholm/Top2}} | ||
| + | ==Nina== | ||
| + | |||
| + | ===Gel clean up=== | ||
| + | |||
| + | I loaded samples in an agarose gel 1 % and ran at 80 V. | ||
| + | |||
| + | Samples: | ||
| + | |||
| + | *Protein A_TAT_N, _Tra10_N and _LMWP_N | ||
| + | *Fusion_CPP1, TAT_C ans _CPP3 | ||
| + | *Fusion_TAT_N, _Tra10_N and _LMWP_N | ||
| + | |||
| + | The procedure was according to the description in protocols. | ||
| + | |||
| + | Weight of all samples: 200 mg | ||
| + | |||
| + | 200 * 3 = 600 ul QXI | ||
| + | |||
| + | In step 3 I added 10 ul of QIAEXII to each sample. | ||
| + | |||
| + | ===Polyacrylamide gel=== | ||
| + | |||
| + | I ran a gel of the purified SOD.His (N terminal) to check if there had been any purification of the protein. | ||
| + | |||
| + | Ladder: PageRuler Unst. Protein Ladder | ||
| + | |||
| + | Arrangement on the gel: | ||
| + | |||
| + | [[Image:Aq18.jpg]] | ||
| + | |||
| + | Gel: | ||
| + | |||
| + | PICTURE!! | ||
| + | |||
| + | The gel was incubated in coomassie blue staining and destained on shake. | ||
| + | |||
| + | Unfortunately there were no bands on the gel, which must mean that there was too small culture (12 ml) and that the cells have not been lysed properly. I will redo this work but with more start culture and lyse the cells in a more effiecient way. | ||
| + | |||
| + | |||
| + | The polyacrylamide gel was prepared by the SDS-PAGE mixtures described in protocols. | ||
| + | |||
==Andreas== | ==Andreas== | ||
===Removal of insertion in BioBrick suffixes=== | ===Removal of insertion in BioBrick suffixes=== | ||
Revision as of 13:47, 25 October 2010
Contents |
Nina
Gel clean up
I loaded samples in an agarose gel 1 % and ran at 80 V.
Samples:
- Protein A_TAT_N, _Tra10_N and _LMWP_N
- Fusion_CPP1, TAT_C ans _CPP3
- Fusion_TAT_N, _Tra10_N and _LMWP_N
The procedure was according to the description in protocols.
Weight of all samples: 200 mg
200 * 3 = 600 ul QXI
In step 3 I added 10 ul of QIAEXII to each sample.
Polyacrylamide gel
I ran a gel of the purified SOD.His (N terminal) to check if there had been any purification of the protein.
Ladder: PageRuler Unst. Protein Ladder
Arrangement on the gel:
Gel:
PICTURE!!
The gel was incubated in coomassie blue staining and destained on shake.
Unfortunately there were no bands on the gel, which must mean that there was too small culture (12 ml) and that the cells have not been lysed properly. I will redo this work but with more start culture and lyse the cells in a more effiecient way.
The polyacrylamide gel was prepared by the SDS-PAGE mixtures described in protocols.
Andreas
Removal of insertion in BioBrick suffixes
Transformation results
Good colony yield on all plates. A few red colonies discovered on all plates, indicating that some partly digested pSB1C3 vector, still containing the RFP insert, had been excised during gel extraction.
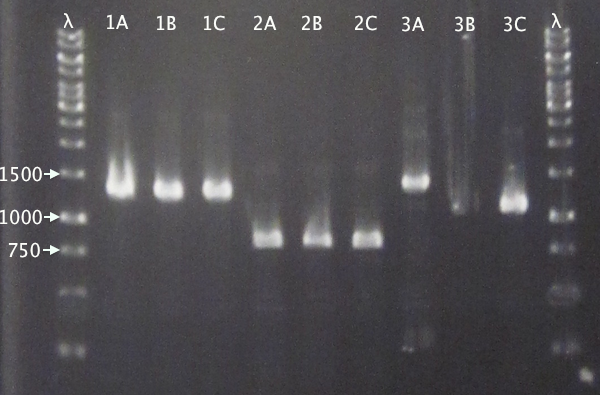
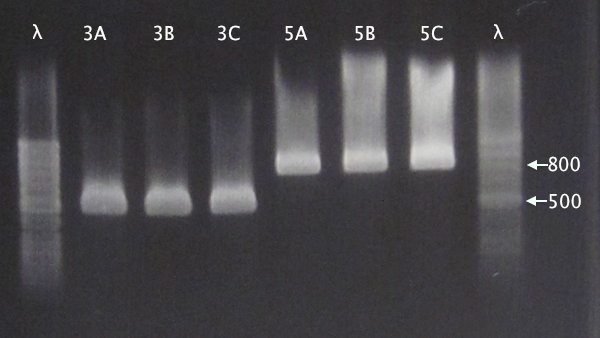
Colony PCR
From 11/10 transformations'
- pSB1C3.IgGp A-C
- pSB1C3.bFGF A-C
- pSB1C3.ProtA A-C
- pSB1C3.yCCS A-C
- pSB1C3.SOD A-C
- Standard colony PCR settings
- 1:15 elongation time
Gel verification
Gel 1: 1 % agarose, 140 V
Gel 2: 1 % agarose, 160 V
Expected bands
- pSB1C3.IgGp: 1262 bp
- pSB1C3.bFGF: 794 bp
- pSB1C3.ProtA: 506 bp
- pSB1C3.yCCS: 1076 bp
- pSB1C3.SOD: 791 bp
Results
- All clones correct.
- All clones correct.
- All clones correct.
- Correct size for clone C, weak band for clone B. Clone A probably carrying an RFP insert.
- All clones correct.
ON cultures
- 5 ml LB + Cm 25; 37 °C, 225 rpm
- 1C, 2C, 3C, 4C, 5C
BL21 transformation and growth test
Testing BL21 cells with/without CPP expression by transforming and inducing with IPTG.
- 50 μl 0.1 M IPTG
- 40 μl competent cells
- pEX.SOD⋅His
- pEX.nTra10⋅SOD⋅His
- pEX.nTAT⋅SOD⋅His
- pEX.nLMWP⋅SOD⋅His
 "
"