Team:Heidelberg/Notebook/miRNA Kit/August
From 2010.igem.org
Laura Nadine (Talk | contribs) |
|||
| Line 2: | Line 2: | ||
{{:Team:Heidelberg/Pagetop|note_mirna_kit}} | {{:Team:Heidelberg/Pagetop|note_mirna_kit}} | ||
= 21/08/2010 = | = 21/08/2010 = | ||
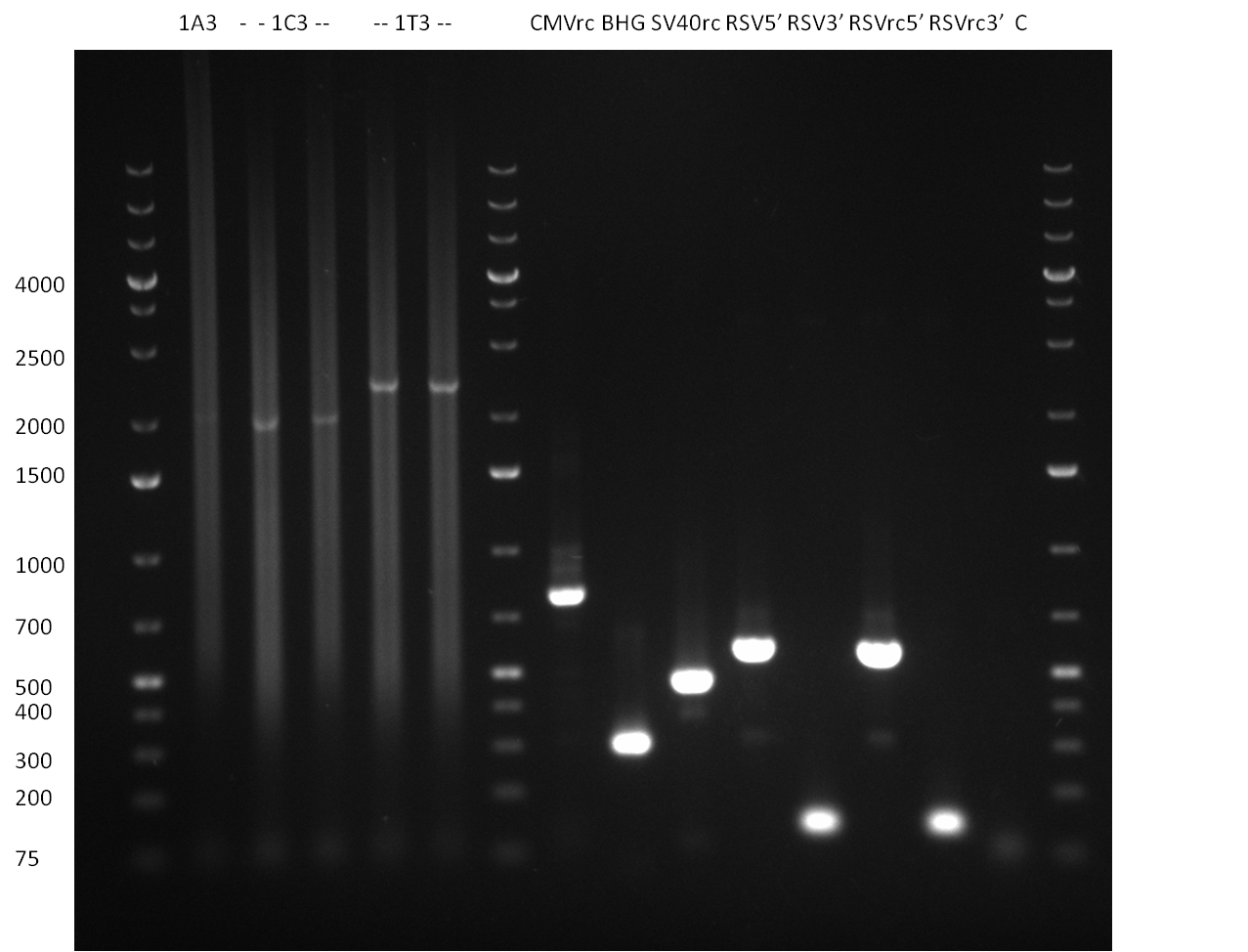
| - | [[Image:20100820_parts.png|thumb| | + | [[Image:20100820_parts.png|thumb|350 px|right|Gel1]] |
| - | [[Image:20100822_gel1.png|thumb| | + | [[Image:20100822_gel1.png|thumb|350 px|right|Gel2]] |
<br /> | <br /> | ||
* all primers for the synthetic miR construction Kit were diluted to a 100 uM concentration | * all primers for the synthetic miR construction Kit were diluted to a 100 uM concentration | ||
| Line 76: | Line 76: | ||
= 22/08/2010 = | = 22/08/2010 = | ||
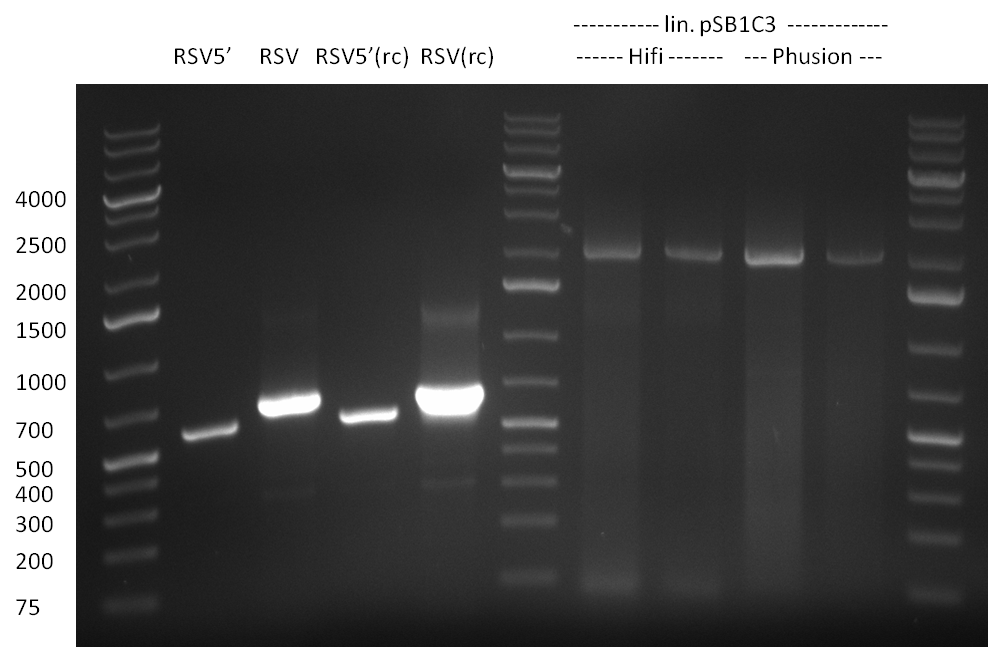
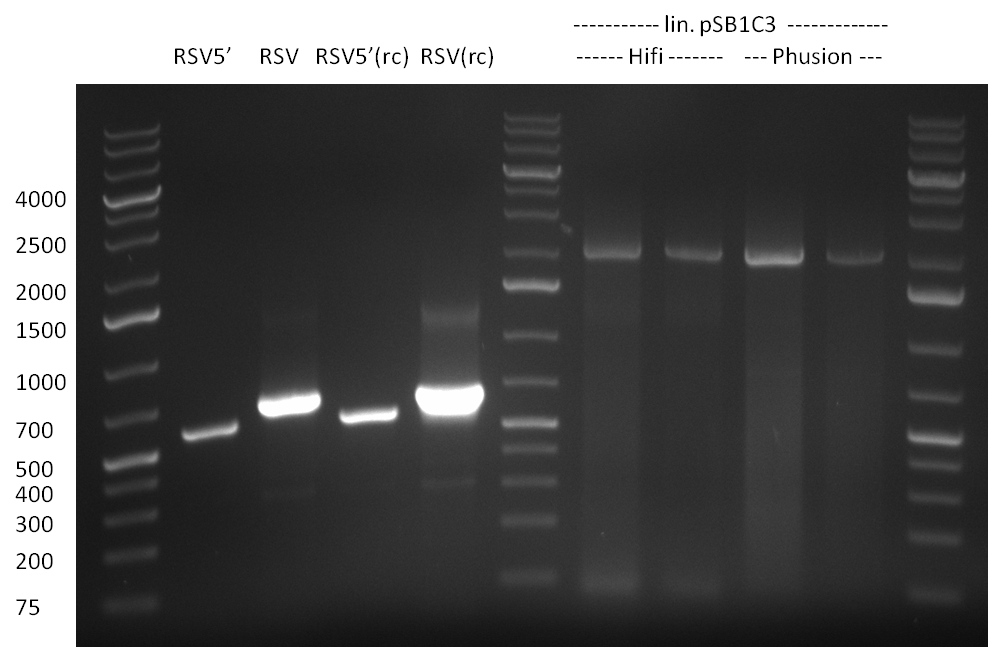
| - | [[Image:20100822_gel2.png|thumb| | + | [[Image:20100822_gel2.png|thumb|350px|right]] |
<br /> | <br /> | ||
* Fusion-PCR for the construction of the whole RSV promoter fragment with BBB prefix and suffix: | * Fusion-PCR for the construction of the whole RSV promoter fragment with BBB prefix and suffix: | ||
| Line 132: | Line 132: | ||
= 23/08/2010 = | = 23/08/2010 = | ||
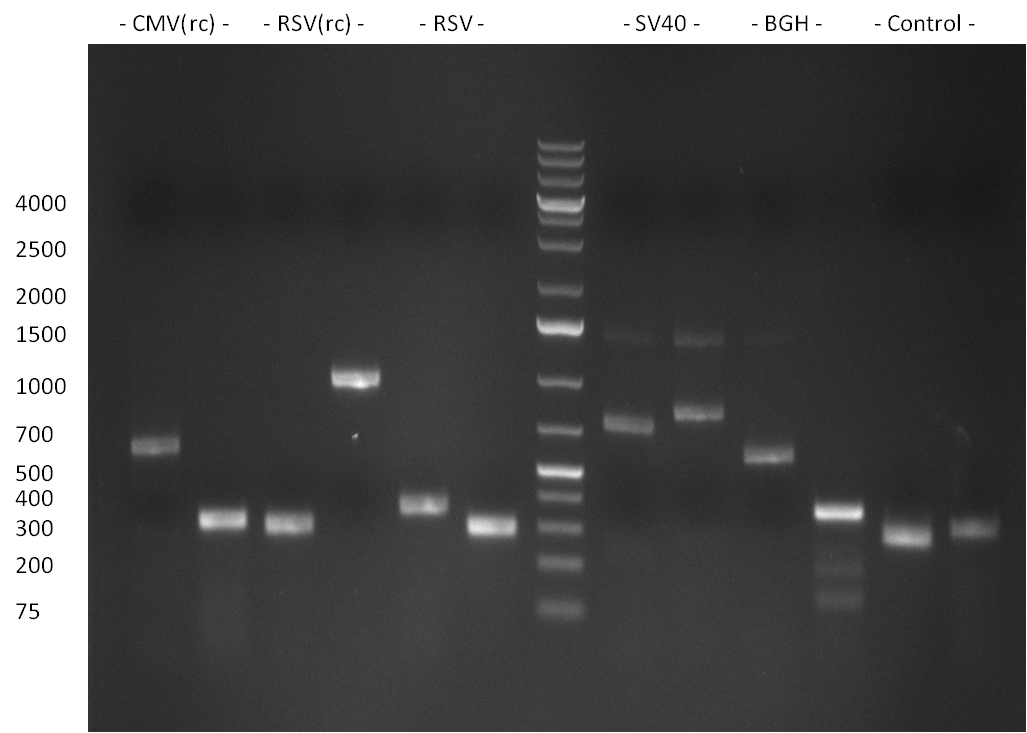
| - | [[image:20100824_gel1.png|thumb| | + | [[image:20100824_gel1.png|thumb|350px|right|gel1]] |
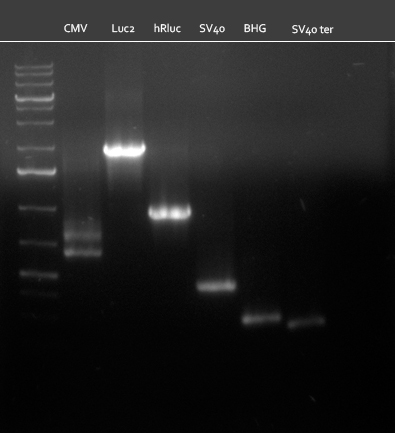
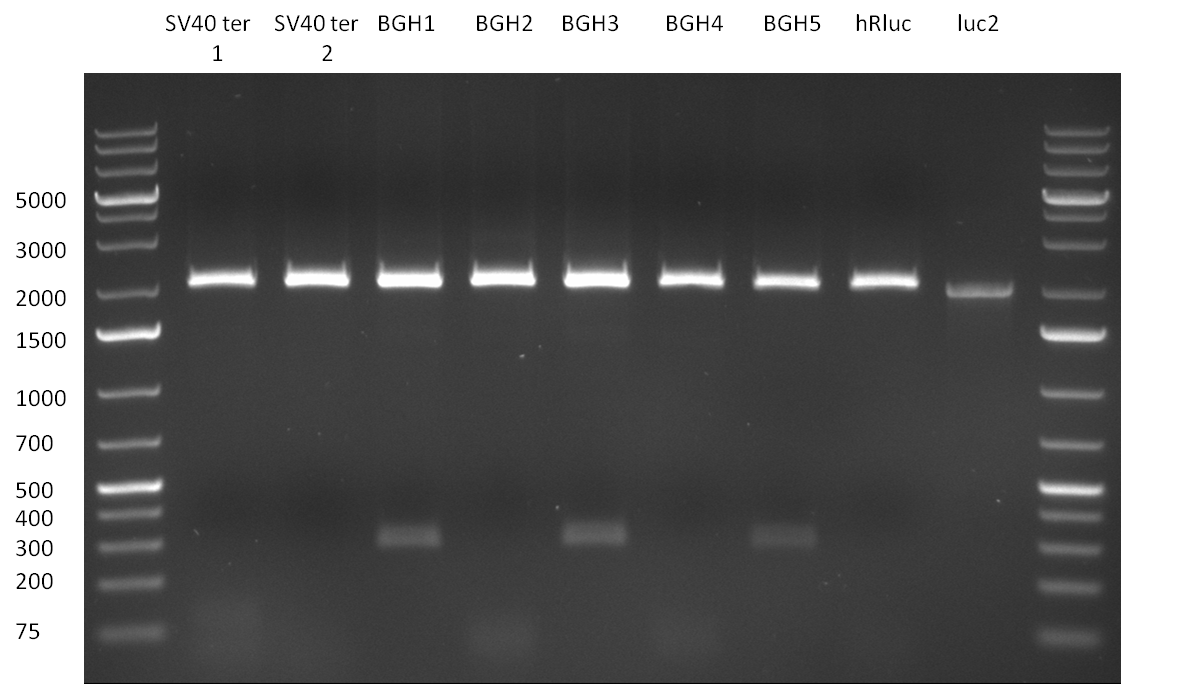
| - | [[Image:20100823_CMV_Luc2_hRluc_SV40_BGH_SV40ter.jpg|thumb| | + | [[Image:20100823_CMV_Luc2_hRluc_SV40_BGH_SV40ter.jpg|thumb|350px|right|Gel2]] |
<br /> | <br /> | ||
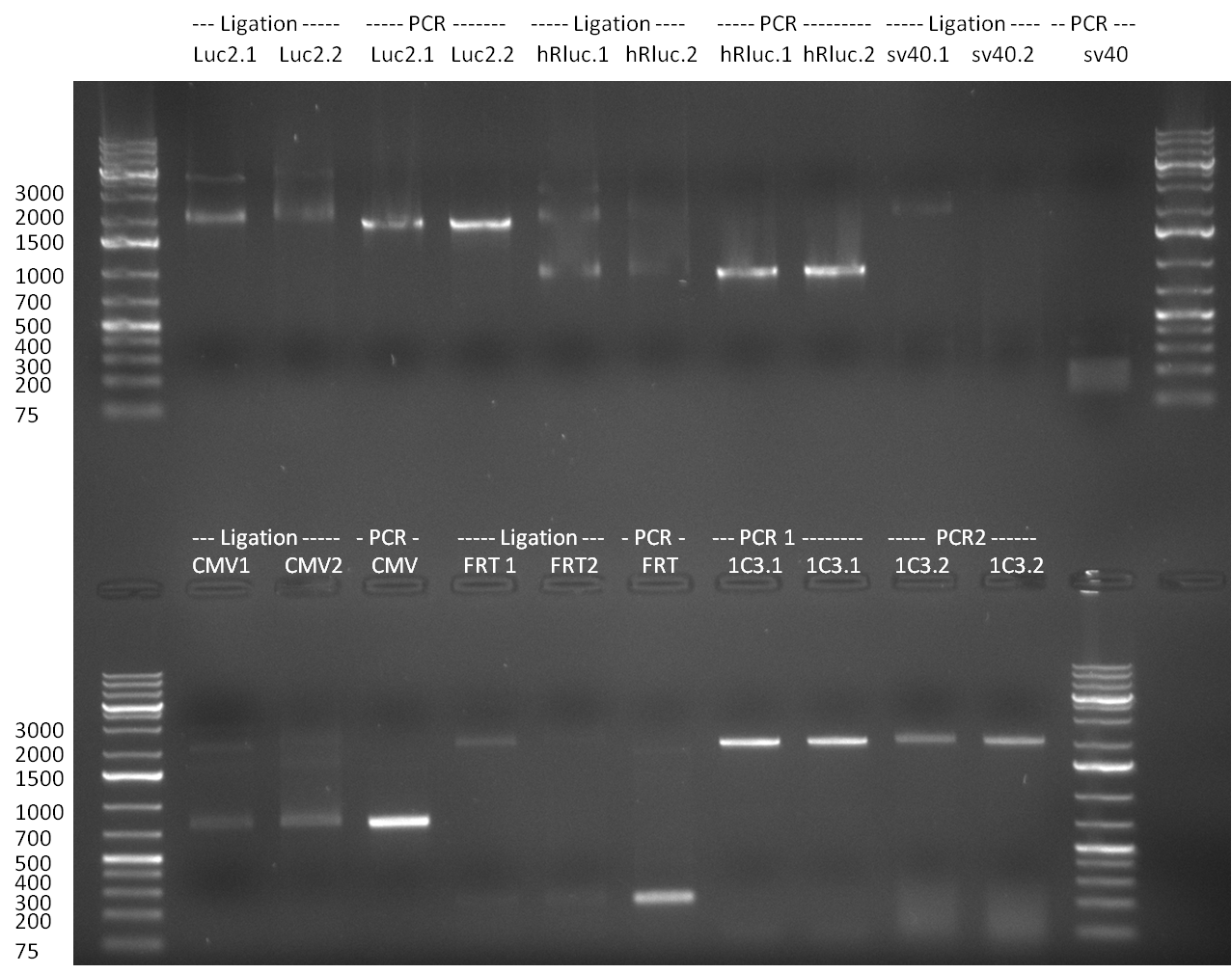
Colony-PCR of the colonies obtained on the agar-plates (cloning previous day) was performed according to the following protocol: | Colony-PCR of the colonies obtained on the agar-plates (cloning previous day) was performed according to the following protocol: | ||
| Line 183: | Line 183: | ||
= 24/08/2010 = | = 24/08/2010 = | ||
| - | [[image: 20100824_gel1_2.png|thumb| | + | [[image: 20100824_gel1_2.png|thumb|350 px|right|Gel1]] |
| - | [[image: 20100824_gel2.png|thumb| | + | [[image: 20100824_gel2.png|thumb|350 px|right|Gel2]] |
| - | [[image: 20100824_gel3.png|thumb| | + | [[image: 20100824_gel3.png|thumb|350 px|right|Gel3]] |
<br /> | <br /> | ||
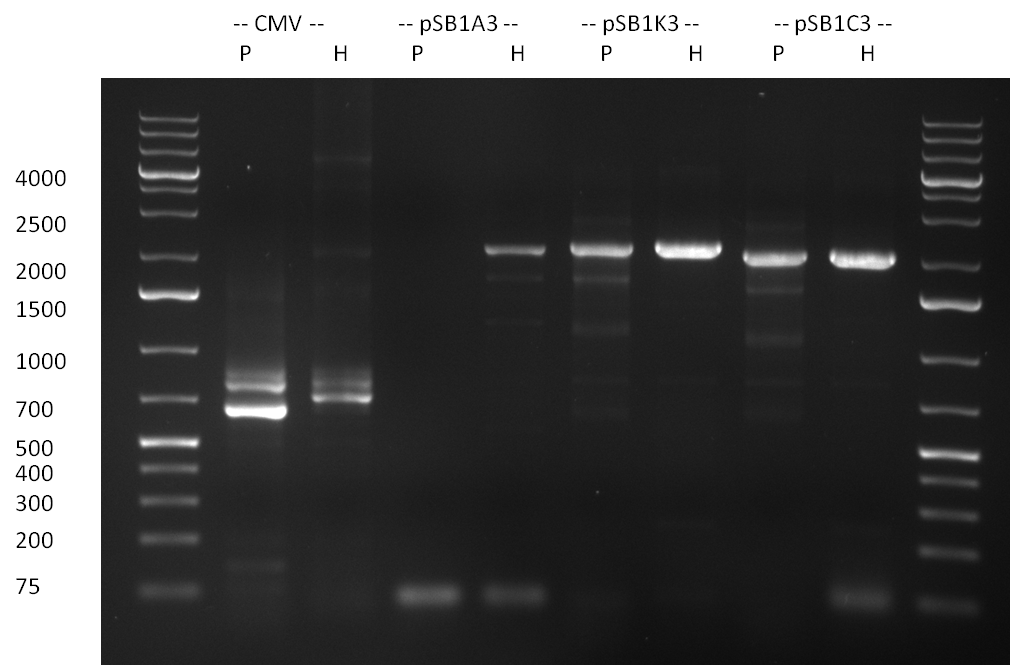
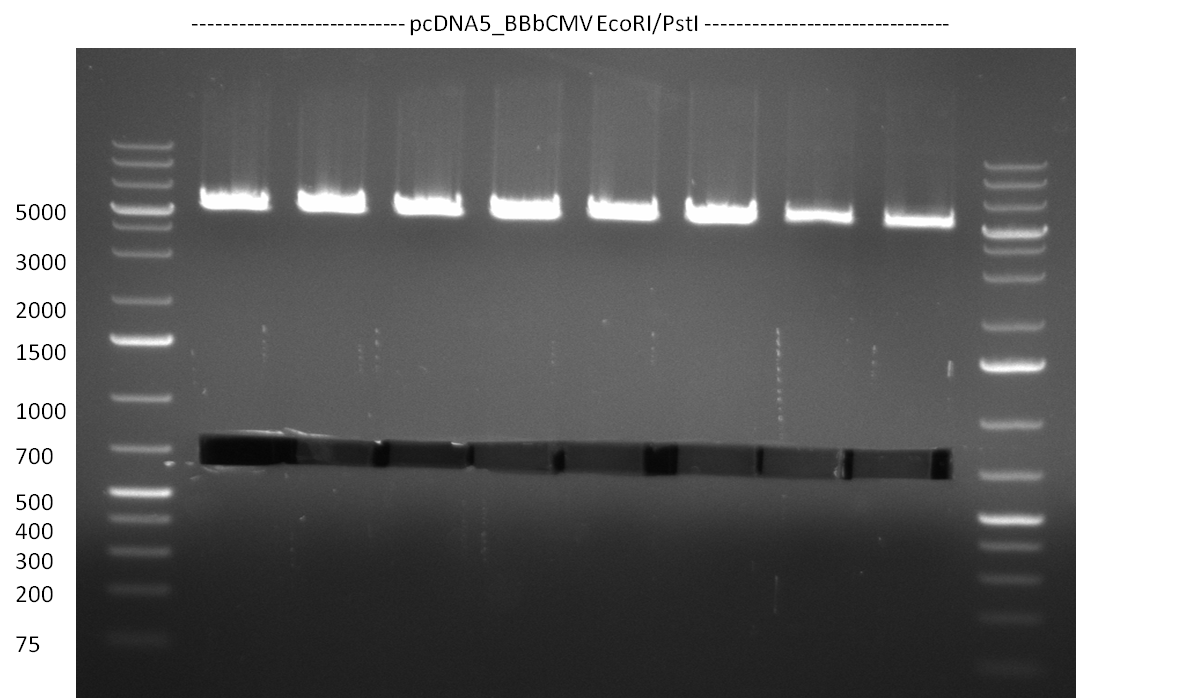
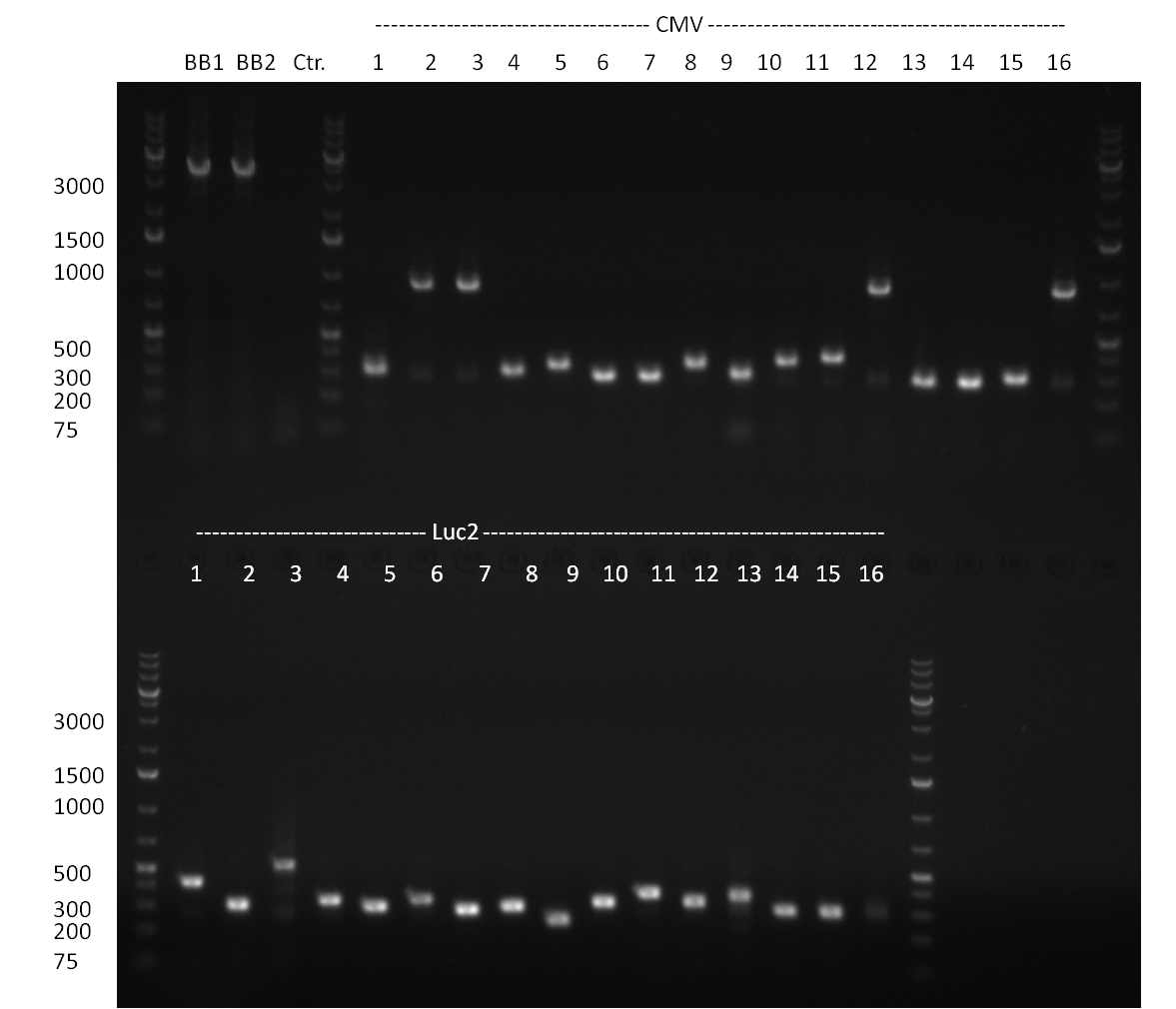
* Amplification of the CMV Promoter as well as the amplification of pSB1A3/C3/K3 standard plasmid backbones was performed via Phusion or Hifi-PCR according to the standard protocol (annealing temperature 61 °C). | * Amplification of the CMV Promoter as well as the amplification of pSB1A3/C3/K3 standard plasmid backbones was performed via Phusion or Hifi-PCR according to the standard protocol (annealing temperature 61 °C). | ||
| Line 249: | Line 249: | ||
<br /> | <br /> | ||
= 27/08/2010 = | = 27/08/2010 = | ||
| - | [[Image:20100827_tet_colonyPCR.png |thumb| | + | [[Image:20100827_tet_colonyPCR.png |thumb|350px|right|gel1]] |
| - | [[Image:20100827_3vectors_digested_EcoPstDpn.png |thumb| | + | [[Image:20100827_3vectors_digested_EcoPstDpn.png |thumb|350px|right|gel2]] |
| Line 310: | Line 310: | ||
= 28/08/2010 = | = 28/08/2010 = | ||
| - | [[Image:20100828_gel1.png|thumb| | + | [[Image:20100828_gel1.png|thumb|350 px|right]] |
| - | [[Image:20100828_gel2.png|thumb| | + | [[Image:20100828_gel2.png|thumb|350 px|right]] |
<br /> | <br /> | ||
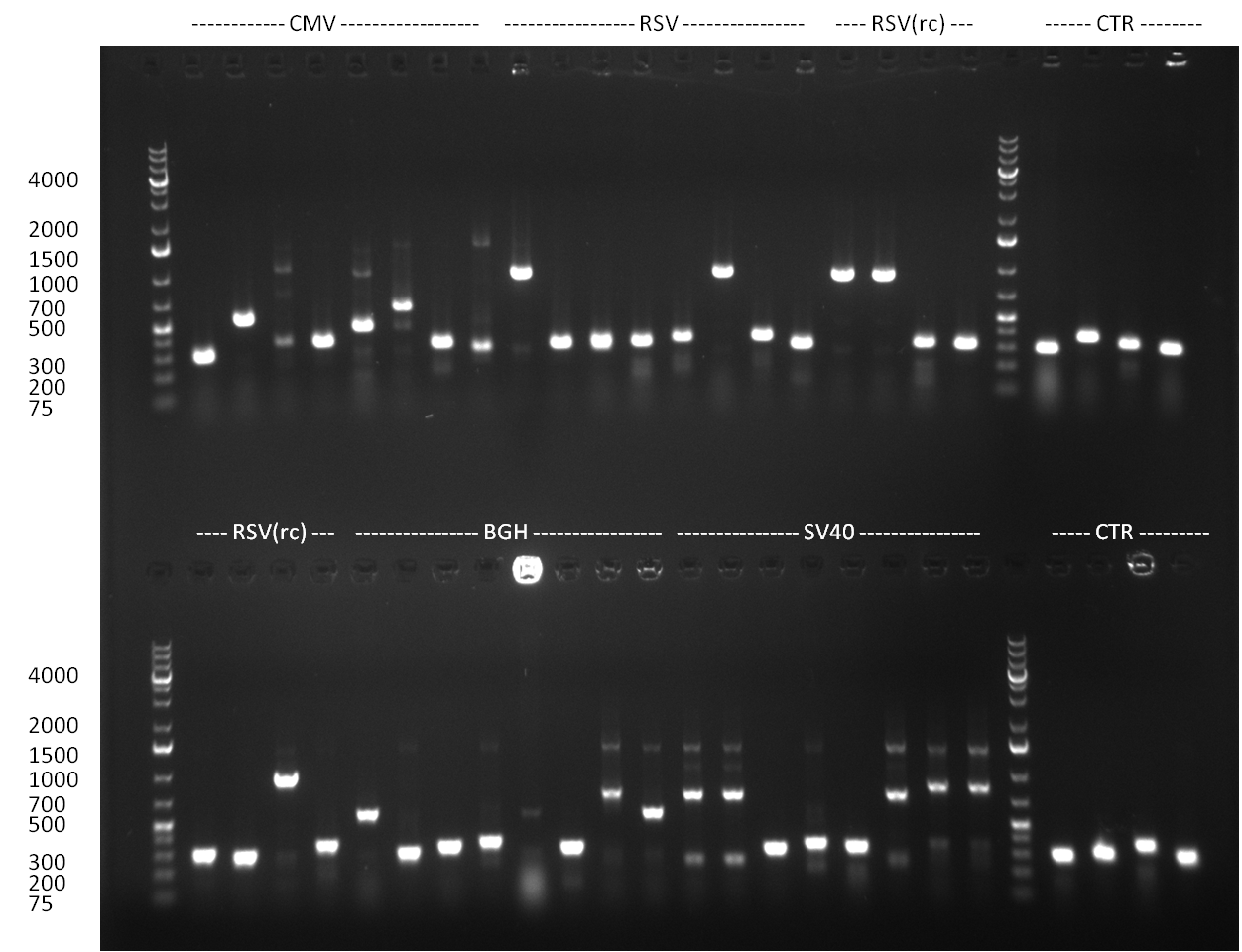
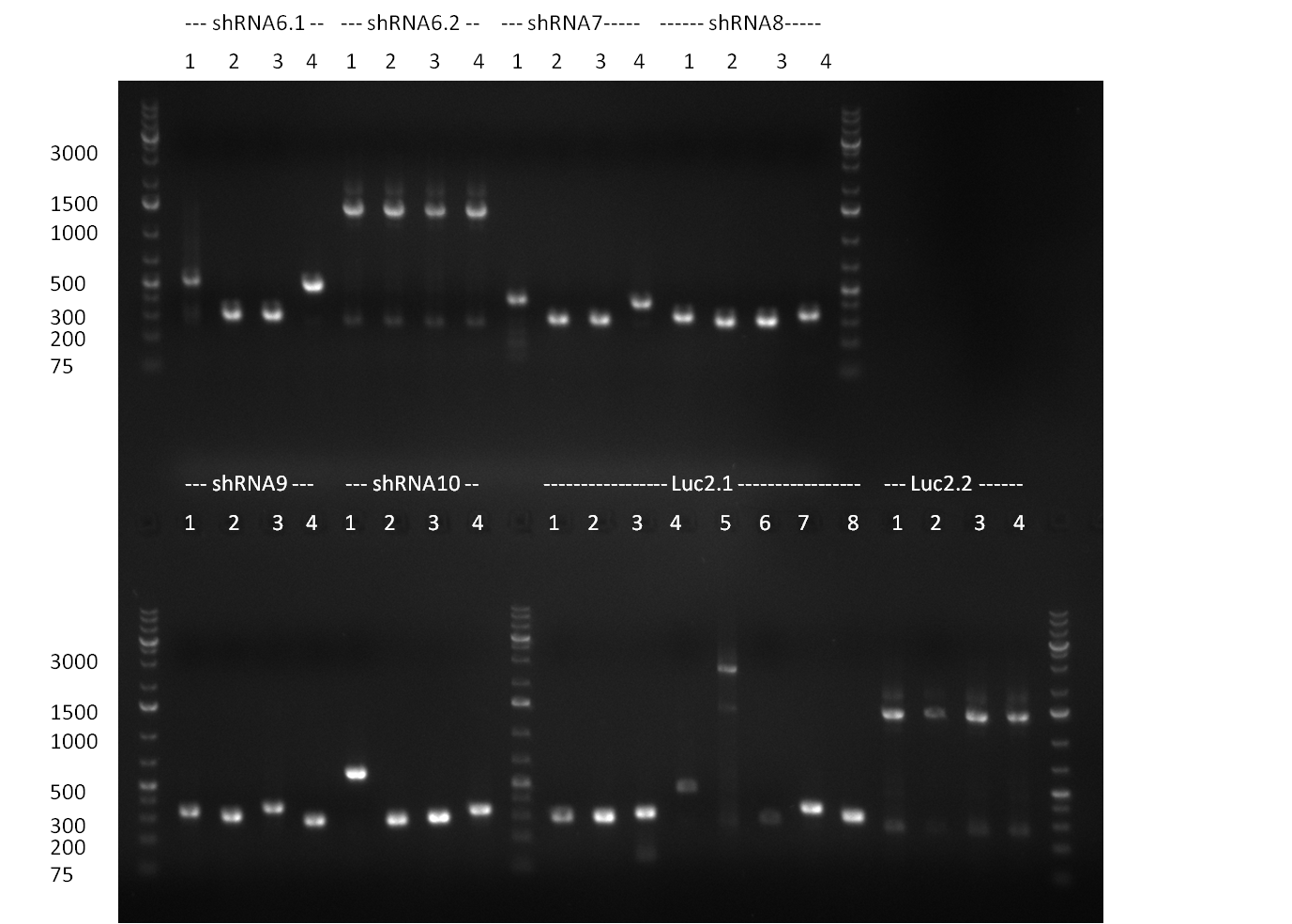
*6 more colonies from the cloning done on the 25th/26th were picked and colony PCR was performed according to the standard colony-PCR protocol. In parallel, Minipreps of each colony were inocculated. The results of the colony-PCR ist shown in the gelpictures. | *6 more colonies from the cloning done on the 25th/26th were picked and colony PCR was performed according to the standard colony-PCR protocol. In parallel, Minipreps of each colony were inocculated. The results of the colony-PCR ist shown in the gelpictures. | ||
| Line 320: | Line 320: | ||
= 29/08/2010 = | = 29/08/2010 = | ||
| - | [[Image:20100829_gel2.png|thumb| | + | [[Image:20100829_gel2.png|thumb|350px|right|gel1]] |
| - | [[Image:20100829_gel1.png|thumb| | + | [[Image:20100829_gel1.png|thumb|350px|right|gel2]] |
<br /> | <br /> | ||
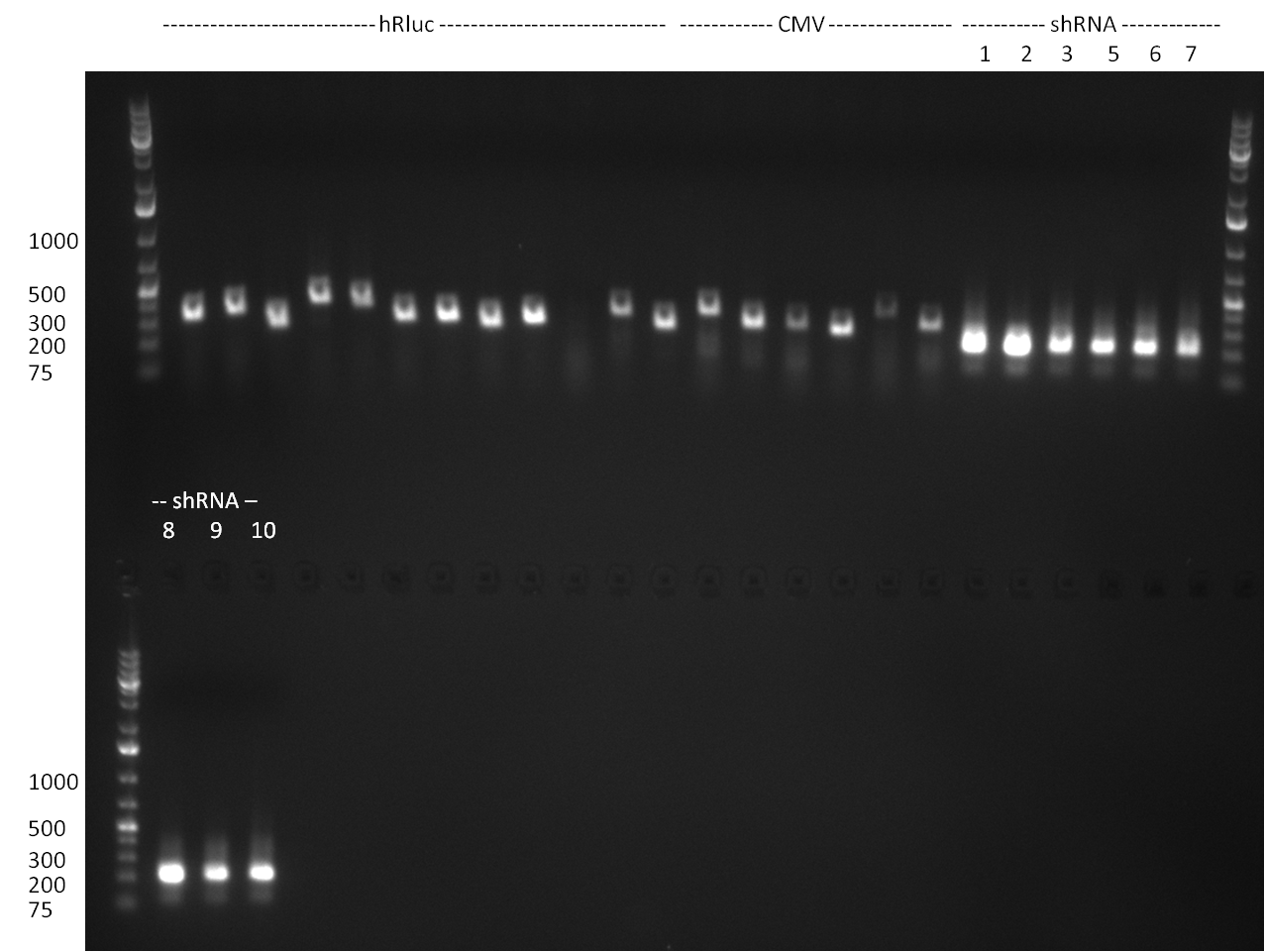
* Vector pcDNA_BBbCMV, PCR product Luc2_ter and shRNA(1-10) with EcoRI/PstI (1 ug each). The BBb_CMV was cut out from the gel (~ 700 bp band) and gel purified (gel1). The PCR product Luc2_ter was purified by applying a nucleotide removal kit. | * Vector pcDNA_BBbCMV, PCR product Luc2_ter and shRNA(1-10) with EcoRI/PstI (1 ug each). The BBb_CMV was cut out from the gel (~ 700 bp band) and gel purified (gel1). The PCR product Luc2_ter was purified by applying a nucleotide removal kit. | ||
| Line 329: | Line 329: | ||
= 30/08/2010 = | = 30/08/2010 = | ||
| - | [[Image:20100830_gel1.png|thumb| | + | [[Image:20100830_gel1.png|thumb|350 px|right|Gel1]] |
| - | [[Image:20100830_gel2.png|thumb| | + | [[Image:20100830_gel2.png|thumb|350 px|right|Gel2]] |
| - | [[Image:20100830_gel3.png|thumb| | + | [[Image:20100830_gel3.png|thumb|350 px|left|Gel3]] |
<br /> | <br /> | ||
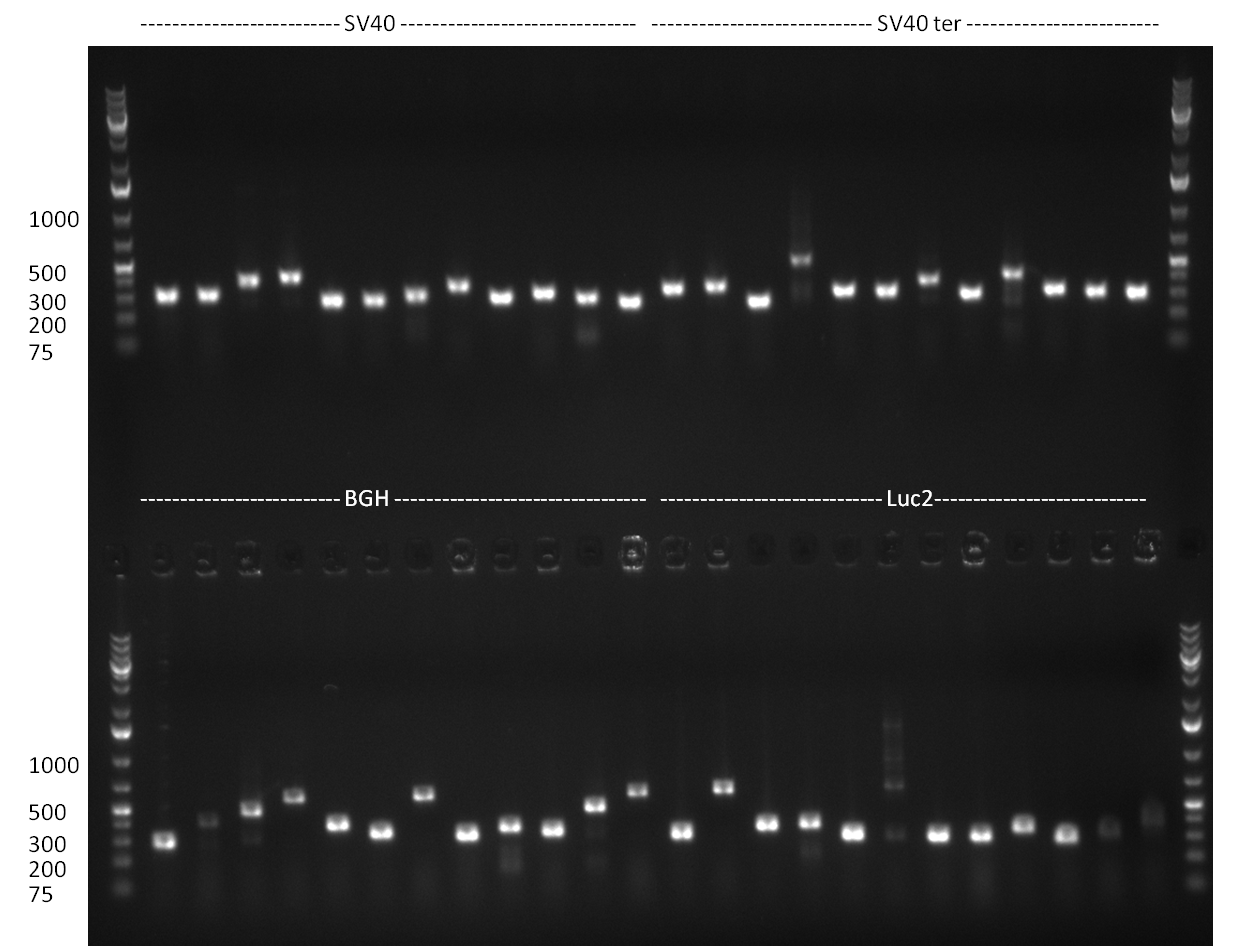
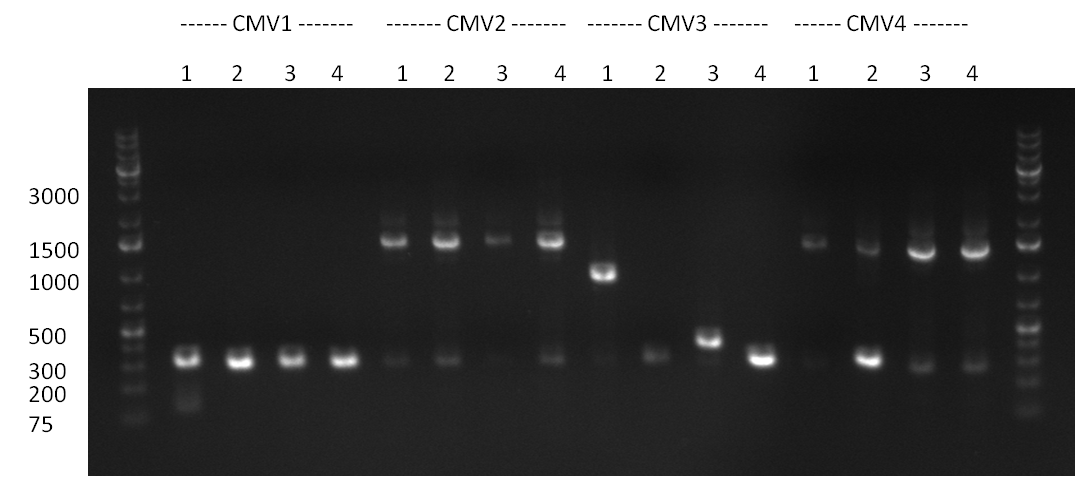
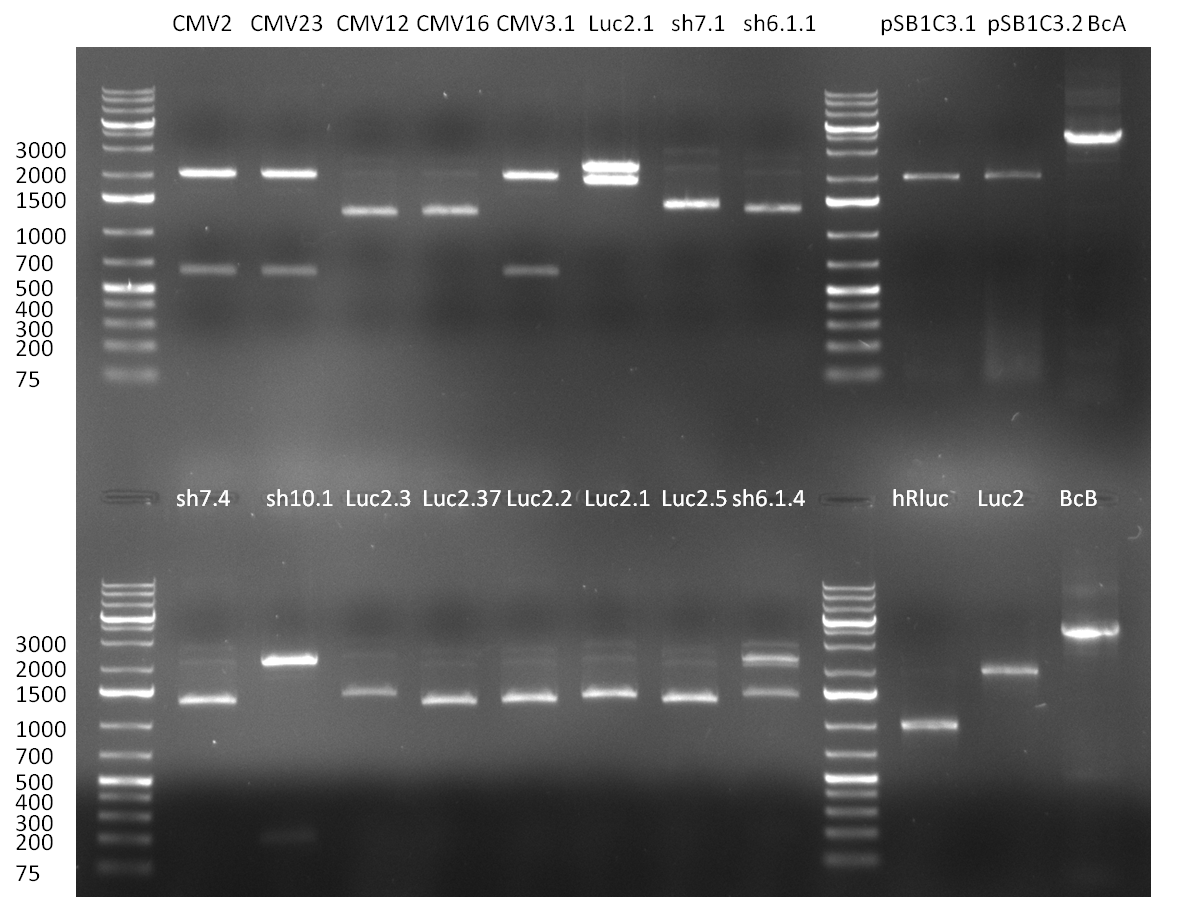
* The ligation result from the previous days' cloning was analyzed via colony PCR (gel 1-3) and all colonies were inocculated (LB cultures) The construct numbers indicate the number of the clone picked from the agar plate. If cloning was performed in Replicates, the first number indicates the plate (i.e. CMV3) and the second number the clone. As seen in gelpicture 1 the clone CMV3.1 and on gelpicture 3 the clones CMV 2,3,12 and 16 all show the right band length of ~1000 bp. On gelpicture 2 the shRNA10.1 clone shows the right band as well as the Luc2.1.5 clone for the Luc2 luciferase. | * The ligation result from the previous days' cloning was analyzed via colony PCR (gel 1-3) and all colonies were inocculated (LB cultures) The construct numbers indicate the number of the clone picked from the agar plate. If cloning was performed in Replicates, the first number indicates the plate (i.e. CMV3) and the second number the clone. As seen in gelpicture 1 the clone CMV3.1 and on gelpicture 3 the clones CMV 2,3,12 and 16 all show the right band length of ~1000 bp. On gelpicture 2 the shRNA10.1 clone shows the right band as well as the Luc2.1.5 clone for the Luc2 luciferase. | ||
Revision as of 10:09, 25 October 2010

|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"