Team:Nevada/Notebook
From 2010.igem.org
(→Team Nevada Notebook) |
(→Team Nevada Notebook) |
||
| Line 525: | Line 525: | ||
** PCR of DREB1C with custom primers using extaq instead of platinum taq | ** PCR of DREB1C with custom primers using extaq instead of platinum taq | ||
** Ran gel of PCR product - Bands showed at 500 bp | ** Ran gel of PCR product - Bands showed at 500 bp | ||
| - | <img src='https://static.igem.org/mediawiki/2010/3/3f/DREB1C_gel_1.jpg'> | + | <html><img src='https://static.igem.org/mediawiki/2010/3/3f/DREB1C_gel_1.jpg'/><html> |
Revision as of 01:23, 25 October 2010
Team Nevada Notebook
Week of April 11-17:
- Christian
- Transformed pBIB into Top 10 Cells
- Bryson, Michael, Senny, Tyler
- Made tobacco cell (NT-1) media in Dr. Shintani's lab
Week of April 18-24:
- Bryson, Christian
- EcoRI digest of pBIB
- Made Na acetate buffer
- Christian
- Ran gel of EcoR1 Digested pBIB
Week of April 25-May 1:
- Bryson
- Ran agarose gel of EcoRI digest
- Matthew
- Team collaborated on pBIB assignment. Too many chefs in the kitchen.
- We decide to each tackle the pBIB problem in parallel
Week of May 2-8:
- Elaine
- Ran 0.8% agarose gel of EcoRI digest
- Made LB/KAN plates
- Spread for colonies
- Did miniprep for pBIB liquid cultures
- Matthew
- Miniprepped pBIB
- Ran EcoRI digest and Klenow
- Ethanol precipitated and ligated
Week of May 9-15:
- Christian
- EcoR1/Xba1 Digest of pBIB
- Did Mini prep on 4 cultures
- Bryson
- Ran 0.8% agarose gel of pBIB post-phenol:chloroform cleanup.
- Did minipreps on additional pBIB liquid cultures.
- Elaine
- Did XbaI and EcoRI digest of pBIB
- Ran 2 0.8% gels of each digest
- Did a miniprep and a Phenol:chloroform clean-up
- Ran 0.8% gel of the XbaI and EcoRI digest with the uncut pBIB
- Matthew
- Ligated and transformed
- Screened colonies, miniprepped, and digested, ran on gel
- No candidates had EcoRI eliminated
Week of May 16-22:
- Christian
- Generated glycerol stock of pBIB transformed Top 10 cells
- Inoculated 5 cultures of pBIB transformed Top 10 cells grown on Kan resistant plates
- Ran samples on gel after digest with EcoR1
- Bryson
- Klenow reactions of EcoRI digests
- Phenol:chloroform cleanup of pBIB prior to ligation
- Blunt-end ligation of klenowed pBIB
- Randy Pares and Vidim Gladwell
- Designed primers pBIB-RB-F, pBIB-RB-R, NOS 3'-Foward Nos3' reverse for sequencing the pBIB plant plasmid.
- Elaine
- Made LB/KAN plates
- Made 50mg/ml stock of KAN
- Made 1X TAE buffer
- Matthew
- Digested and tested more colonies, none worked
- Started over with pBIB and Klenow
- Ethanol precipitated and ligated
- Transformed
- Screened colonies, miniprepped, and digested, ran on gel
- No candidates had EcoRI eliminated
Week of May 23-29:
- Christian
- Miniprep on pBIB transformed Top 10 cells using "low copy #" modification
- Digested with EcoR1 and HindIII
- Bryson
- Incubated 50 mL liquid culture of E. coli with pBIB (samples 3).
- Miniprepped sample 3, using modified protocol for large/low-copy plasmids.
- EcoRI digest of uncut sample 3
- Prepared 5 mM dNTP stock
- Elaine
- Incubated 50 mL liquid culture of E. coli with pBIB (sample 4 & sample 5)
- Qiaprep miniprep of sample 4 & sample 5 according to manufacturer's modified protocol for large/low-copy plasmids
- Nanodrop of DNA recovery of miniprepped sample 4 & sample 5
- Randy and Vadim
- Sent pBIB vector to Nevada Genomics Center for sequencing with primers designed during the week of May 16-22
- Designed forward and reverse minimal ccdB primers for PCR and sequencing of ccdB gene
- Maxiprepped pBIB vector using QIAGEN QIAfilter Plasmid Maxi Kit
- Matthew
- Investigated alternative ways of eliminating EcoRI site.
- Want to try hexameric linkers
Week of May 30-June 5:
- Christian
- Miniprep Top 10 Cells transformed with pBIB
- Bryson
- HinD3 digest of uncut sample 3
- Ran 0.8% gels of samples 1-5 to verify complete digestion by EcoRI
- Pooled uncut samples 3, 4 and 5 (pBIB-pool)
- Recieved 5 µg of pBIB from Randy and Vadim's maxi-prep (pBIB-maxi)
- EcoRI digest of pBIB-pool and pBIB-maxi
- Ran 0.8% agarose gel of EcoRI digests
- Klenow reactions of pBIB-pool and pBIB-maxi
- Made glycerol stocks of pBIB samples 1-5
- Elaine
- EcoRI digest of uncut pBIB sample 4 and 5
- HinD3 digest of uncut sample 4 & sample 5 as a positive control
- Ran 0.8% gels of samples 1-5 to see if EcoRI digest was successful
- Ethanol precipitation of the EcoRI digests of sample 4 & sample 5
- Nanodrop resulted to a low DNA content
- Worked with Bryson for the EcoRI digest of the 5µg of pBIB maxi-prep
- Ran 0.8% agarose gel of the pBIB maxi-prep EcoRI digest
- Modified all protocols of the Binary vector
- Randy and Vadim
- Calculated amount of reagent needed for Deep Vent DNA polymerase reaction (New England Biolabs)
- Programmed thermal cycler for PCR of ccdB gene
- Ran PCR for ccdB
- Prepared LB/KAN Broth
- Gel analyzed resultant ccdB PCR reaction with 1.2% agarose gel
- Reaction was unsuccessful
- Matthew
- Miniprepped pBIB
- Ran EcoRI digests
- Ran ethanol precipitation
- Ligated with several concentrations of hexameric linkers designed to destroy site
- Ran transformation, no colonies
- Believed procedures/conditions ran on ligation and transformation not ideal
Week of June 6-12:
- Christian
- Klenow reaction on EcoR1 digested pBIB
- T4 ligation on Klenow products
- Bryson
- Ligation reactions for pBIB-pool and pBIB-maxi
- Digested pBIB-pool and pBIB-maxi again with EcoRI to linearize any unmodified pBIB
- Transformed Top-10 Cells with modified pBIB (designated pBIB#)
- Obtained two colonies after overnight incubation
- Line-streaked the two colonies (pBIB# 1 and pBIB# 2) on an LB-Kan plate
- Randy and Vadim
- Uploaded pBI101, pBIN19, pBIB-KAN, and ccdB gene to Vector NTI
- Ordered second set of pBIB primers: pBIB-RB-F2, pBIB-RB-F3, pBIB-RB-R2, and pBIB-RB-R3 for sequencing of pBIB
- Modified thermal cycler conditions for PCR of ccdB gene
- Ran PCR for ccdB using HiFi DNA polymerase (Invitrogen)
- Gel analyzed resultant ccdB PCR reaction with 1.2% agarose gel and successfully amplified ccdB gene
- Maxiprepped pBIB vector using QIAGEN QIAfilter Plasmid Maxi Kit
- Matthew
- Miniprepped pBIB
- Ran Digestions and ethanol precipitation
- Ligated with hexameric linkers
- Transformed, had some colonies
Week of June 13-19:
- Christian
- EcoR1 digest of ligation product
- Transformed Top 10 Cells with pBIB from ligation and plated
- Inoculated 30 colonies
- UNR mini prep on all 30 colonies
- Digested Samples with HindIII and EcoR1
- QIAGEN mini prep on samples 11,12 &13
- Digested Samples with HindIII and EcoR1
- Bryson
- Prepared liquid cultures of pBIB# 1 and pBIB# 2
- Miniprepped liquid cultures of pBIB#
- EcoRI and HinDIII digests of pBIB#
- Ran agarose gel of pBIB cut and uncut, pBIB# uncut, pBIB# cut with EcoRI and pBIB# cut with HinDIII to confirm the absence of EcoRI site in pBIB#
- Single-colony streaked pBIB# 2 on a fresh LB-Kan plate
- Elaine
- Worked with Chris to incubate 30 liquid cell cultures
- Ran 0.8% gels of all 30 samples
- Randy and Vadim
- Topocloned ccdB gene into TOPO PCR Blunt II vector
- Determined concentration of pBIB maxipreps using PicoGreen analysis
- Single-colony isolated nine colonies of TOPO-cloned ccdB gene
- Miniprepped cultures of ccdB gene in TOPO-clone
- Digested minipreps using EcoRI
- Ran 1% gel for digested minipreps
- Single colony streaked four cell lines of ccdB gene in TOPO-clone: line 3, line 7, line 8, line 9
- Ordered primers for Left Border Repeat of pBIB: pBIB-LB-F and pBIB-LB-R for sequencing of pBIB vector
- Matthew
- Miniprepped candidates and digested them
- Ran on gel, some had strange bands, I want to retest
- Colonies appear to not have eliminated EcoRI
Week of June 20-26:
- Christian
- Ran gel of Sample 11
- Bryson
- Selected 3 colonies from pBIB# 2 (2-1, 2-2, 2-3) and streaked on a fresh LB-Kan plate
- Started 20 mL liquid cultures of 2-1, 2-2, and 2-3
- Miniprepped samples
- EcoRI and XbaI digests of samples to confirm that the EcoRI was gone and 2-1, 2-2, 2-3 arose from the same colony
- 0.8% agarose gel of digests
- Randy and Vadim
- Miniprepped isolated cultures of ccdB gene using QIAGEN QIAprep Spin Miniprep Kit
- Nanodropped minipreps
- Digested minipreps using EcoRI
- Ran 1% gel for digested minipreps
- Sent cell lines 7-2 and 8-2 for sequencing to Nevada Genomics Center with ccdB M13 Forward and Reverse primers added
- Analyzed ccdB samples using Vector NTI (Invitrogen)
- Sent cell lines 7-2, 8-2, and 9-1 for sequencing/resequencing to Nevada Genomics Center with ccdB M13 Forward and Reverse primers added
- Elaine
- Primer Design of RD29A
- Matthew
- Attempted to test more candidates from hexameric ligation, no candidates had eliminated EcoRI
- Team reevaluated its standing, given a month of failed attempts to modify pBIB
- We are each assigned new tasks. I will be put in charge of making EYFP and mCherry with plant Kozak
- I design primers to extract parts from iGEM vectors with Kozak sequences
Week of June 27-July 3:
- Randy and Vadim
- Prepared ccdB for MaxiPrep using QIAGEN QIAfilter Plasmid Maxi Kit
- Prepared thermal cycler protocol for pBIB vector
- Performed multiple PCR on pBIB vector
- Transformed ccdB into NEB cells unsuccessfully
- Made chlorophenocol resistant agar plates
- Agarose gel analyzed PCR products
- Matthew
- Transformed colonies with iGEM parts, miniprepped.
- Attempted PCR on EYFP, Failed. Reevaluated protocol.
Week of July 4-10:
- Randy and Vadim
- Agarose gel analyzed PCR products
- Transformed ccdB into NEB, OmniMax, and DEB cells
- Made Kanamycin resistant LB plates
- Elaine
- Transformed ccdB into NEB, OmniMax, and DEB cells
- Made LB/KAN plates
- Took picture of ccdB Sensitivity Experiment 1 Results
- GFP Primer Design
Week of July 11-17:
- Elaine
- Made LB/Amp plates
- Randy and Vadim
- TOPOcloned pBIB fragment
- Attempted to ligate ccdB into iGEM vector pSB1C3
- Single colony streaked ccdB transformation
- Made Chloramphenicol broth
- Performed PCR on pBIB cell lines
- Digested ccdB in iGEM vector pSB1C3 with PstI and EcoRI
- Miniprepped ccdB colonies
- Gel analyzed pBIB PCR product
- Transformed pSB1C3+RFP vector onto Chloramphenicol resistant plates
- Made 50x TAE buffer
- Miniprepped pSB1C3+RFP vector
- Matthew
- Processed Miniprepped iGEM colonies for mCherry and EYFP
- Set up PCR for mCherry, EYFP, and NOS
Week of July 18-24:
- Elaine
- PCR of GFP
- Ran agarose gel analysis of GFP PCR product
- Randy and Vadim
- Amplified pBIB fragment using PCR
- Gel analyzed pBIB PCR product
- Attempted ligation of ccdB into iGEM vector pSB1C3
- Transformed ligation into NEB cells
- Selected supposed colonies with ccdB+pSB1C3
- Matthew
- Analyzed PCR fragments on gels for mCherry, EYFP, and NOS
PCR results: worked for eyfp and mcherry with bands around 700bp.
From Left: Lane 1 - Lamda Hind III Mlc. Marker.
Lane 2 - Uncut iGem vector
Lane 3 - EYFP
Lane 4 - EYFP
Lane 5 - EYFP
Lane 6 - mCherry
Lane 7 - mCherry
Lane 8 - mCherry
Lane 9 - 100bp ladder
- NOS PCR did not appear to work.
- Performed transformation protocol with TOPO vector
- Performed screening on Kan and miniprepped
Week of July 25-31:
- Christian
- Transformed Top 10 cells with pBIB and pMA (containing rd29A)
- Elaine
- GFP Transformation
- Cell Culture of GFP
- Miniprep of GFP
- EcoR I & Pst Digest of GFP
- Ran 1.2% agarose gel analysis of the GFP digest
- PCR of GFP
- Randy and Vadim
- Miniprepped ccdB+pSB1C3 colonies
- Digested minipreps with EcoRI and PstI
- Performed colony PCR on pSB1C3 colonies
- Matthew
- Ran Digests and gels for candidates of mCherry, EYFP, and NOS in TOPO
- 1 possible candidate each for mChery, EYFP, and NOS.
- Nos bands did not show but sent one for sequencing anyway
Week of August 1-7:
- Christian
- Designed and ordered primers for wt Arabidopsis isolation of DREB1C promoter region
- Mini prep on pBIB and pMA inoculations
- Elaine
- Ran 1.2% Agarose Gel analysis of GFP PCR product
- Topocloned GFP PCR product
- Cell Culture of GFP Topocloned colonies
- Streaked colony plate
- Miniprep of GFP Topocloned colonies
- EcoR I digestion of GFP Topocloned
- Ligation Test: GFP original vector digested with EcoR I
- Samantha and Christian
- pMA and pBIB liquid cultures and mini preps
- Bryson
- Designed and ordered primers for the 35S promoter
- Made spectymycin plates
- Transformed DB3.1 cells with pK7FWG2
- Liquid cultures and minipreps to isolate pK7FWG2
- Randy and Vadim
- Grew ccdB 8-2 on Kan. resistant plates
- Gel analyzed colony PCR product from July 29
- Made additional LB broth
- Gel analyzed ccdB+pSB1C3 ligation using EcoRI and Pst
- Ligation was unsuccessful
- Performed PCR cleanup on ccdB
Week of August 8-14:
- Elaine
- Ran 1.2% Agarose Gel analysis of GFP Topocloned EcoR I digest
- Ligation Test: Ligated EcoR I digest GFP original vector
- Ligation Test: Transformed Ligation and EcoR I digest of GFP into NEB cells
- Cell culture and streaked colonies of RFP in PSB1C3 vector
- Miniprepped RFP in PSB1C3 vector
- Digested both RFP in PSB1C3 vector & GFP Topo Vector together with EcoRI and PstI
- Ethanol Precipitation of Digested RFP & GFP in PSB1C3 & Topo Vectors
- Ligated the RFP & GFP in either PSB1C3 & Topo Vectors
- Transformed RFP & GFP in either PSB1C3 & Topo Vectors by growing in NEB Cells in KAN & Chloramphenicol plates
- Samantha and Christian
- Triple digest of pMA and pBIB with Hind III, Mfe I, and Eco RI
- Ligation of triple digest products
- Bryson
- PCR of pK7FWG2
- Randy and Vadim
- Made and aliquoted ligase buffer, ATP, and DTT for use for ligation
- Ligated ccdB+pSB1C3, did not use EtOH precipitation
- Miniprepped ccdB+pSB1C3 ligation
- Digested and gel analyzed ligation using EcoRI and Pst
- Ligation was unsuccessful
- Ligated ccdB+pSB1C3 using EtOH precipitation
Week of August 15-21:
- Elaine
- Selected 20 colonies for cell culture and streaked into KAN & Chloramphenicol plates
- Miniprepped 11 cell cultures that didn't grow in KAN plates which means it has to be GFP in PSB1C3 vectors
- Digested GFP in PSB1C3 vectors with EcoR1 & PstI
- Ran 1.2% Agarose Gels analysis of GFP in PSB1C3 vectors digested with EcoR1 & PstI
- Bryson
- 1.2% gel to confirm presence of amplicon
- Topocloning of amplicon and transformation of Topo vector into DB3.1 cells
- Randy and Vadim
- Miniprepped ligation from Aug. 13
- Digested and analyzed ccdB+pSB1C3 ligation using EcoRI and Pst
- Ligation was unsuccessful
- Ligated ccdB+pSB1C3 using EtOH precipitation
- Made Chlor. resistant plates
- Autoclaved labware
Week of August 22-28:
- Christian
- Ran PCR on genomic Arabidopsis thaliana DNA using custom primers
- Ran gel of PCR product, no bands corresponding to DREB1C
- Elaine
- Sequenced colony #11 & #17 of GFP in the PSB1C3 vectors (submitted 4: 11F, 11R, 17F, 17R)
- Made chloramphenicol plates and cell cultures from colony #17 & streaked more of colony # 17
- Miniprepped 8 cell cultures and nanodrop
- Ran 1.2% Agarose Gel to check for DNA (GFP in PSB1C3 vector)
- Bryson
- Isolated and streaked 6 colonies
- Liquid cultures and minipreps of samples
- Digest of samples with EcoRI and PstI to confirm presence of insert
- Randy and Vadim
- Miniprepped colonies from ligation from Aug. 20
- Digested and gel-analyzed ccdB+pSB1C3 ligation
- Ligation was unsuccessful
- Ligated ccdB+pSB1C3 using EtOH precipitation
- Made Chlor. resistant plates
Week of August 29-September 4:
- Elaine
- Ran 1.2% Agarose Gel to check for DNA (GFP in PSB1C3 vector)
- Sequencing analysis of colony #11 & #17
- Bryson
- Grew 20 mL liquid cultures of pSB1C3 and Topo vector to prepare for ligation
- Miniprepped cultures and eluted in 300 microliters
- 50 microliter digests done
- 20 microliters were run on 1.0% agarose gel to confirm complete digestion of both plasmids
- Digested plasmids were added in a roughly 1:1 ratio and ligated in a 40 microliter ligation reaction
- Randy and Vadim
- Digested and gel-analyzed ccdB+pSB1C3 ligation from Aug. 27
- Ligation was unsuccessful
- Performed PCR on ccdB
- Ligated ccdB+pSB1C3 using Phenol:Chloroform cleanup techniques and EtOH precipitation
- Digested and gel-analyzed ccdB+pSB1C3 ligation from Aug. 27
- Matthew
- Sequence results from first attempt at mCherry,EYFP, and NOS failed. NOS abandoned.
- Started over with new colonies, miniprepped, digested, ethanol precipitated, and ligated for Topo
- Screened colonies on Kan and Amp, selected several to miniprep
- Digested and ran on gels, identified several more candidates for sequencing for mCherry and EYFP
Week of September 5-11:
- Elaine
- Sequenced 4 tubes of #17 of GFP in PSB1C3
- Digested RD29A with Hind III & MfeI-HF
- Analysis of sequence of GFP (PSB1C3)
- Bryson
- Transformation of ligation reaction
- Plated on chloramphenicol
- Selected four white colonies and single-colony streaked them
- 4 mL liquid cultures of colonies
- Miniprepped samples and eluted in 50 microliters
- 20 microliter digests with EcoRI and PstI done to ensure insert was present in pSB1C3
- Randy and Vadim
- Transformed ccdB+pSB1C3 ligation from Sept. 3 into DB3 cells
- Transformation unsuccessful
- Made Chlor. resistant plates
- Made new 3M NaOH stock
- Transformed ccdB+pSB1C3 ligation from Sept. 3 into DB3 cells
- Matthew
- Miniprepped candidates for mCherry and EYFP in Topo vector
- Ran digests and gels on topo candidates and submitted them for sequencing
- Sequencing Results came back positive for mCherry and EYFP in Topo
- mCherry showed a point mutation but it should not have an effect on translation
Week of September 12-18:
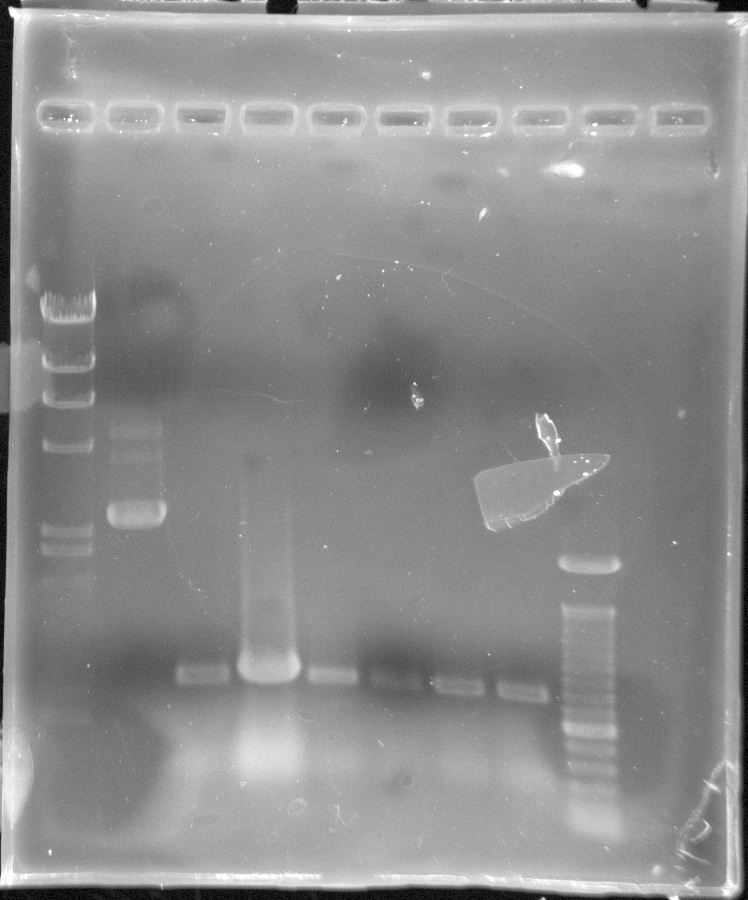
- Christian
- PCR of DREB1C with custom primers using extaq instead of platinum taq
- Ran gel of PCR product - Bands showed at 500 bp
 * ''Elaine''
** RD29A(pMA) with RFP(PSB1C3) EcoR1 & Pst I Digest
** Ethanol Precipitation of digested synthetic RD29A + RFP(pMA) with RFP(PSB1C3)
** Ligation of digested synthetic RD29A + RFP(pMA) with RFP(PSB1C3)
** Transformation of ligated synthetic RD29A + RFP(pMA) with RFP(PSB1C3)
* ''Randy and Vadim''
** Miniprepped ccdB with Mfe site in TOPO vector
** Miniprepped RFP in pSB1C3
** Digested and gel analyzed ccdB with Mfe samples using EcoRI and Pst
** Digested ccdB in TOPO vector and RFP in pSB1C3 using EcoRI and Pst
** Gel purified ccdB and pSB1C3 fragments from digested samples
* Samantha
** Gel extraction of RD29A
* ''Matthew''
** Miniprepped, digested, and ran on gels candidates for mCherry and EYFP with Kozak
** Several candidates possible for sequencing for EYFP and 3 candidates for mCherry
[[Image: Polasko_EYFPigem.jpg|300px|thumb|left]]
* ''Elaine''
** RD29A(pMA) with RFP(PSB1C3) EcoR1 & Pst I Digest
** Ethanol Precipitation of digested synthetic RD29A + RFP(pMA) with RFP(PSB1C3)
** Ligation of digested synthetic RD29A + RFP(pMA) with RFP(PSB1C3)
** Transformation of ligated synthetic RD29A + RFP(pMA) with RFP(PSB1C3)
* ''Randy and Vadim''
** Miniprepped ccdB with Mfe site in TOPO vector
** Miniprepped RFP in pSB1C3
** Digested and gel analyzed ccdB with Mfe samples using EcoRI and Pst
** Digested ccdB in TOPO vector and RFP in pSB1C3 using EcoRI and Pst
** Gel purified ccdB and pSB1C3 fragments from digested samples
* Samantha
** Gel extraction of RD29A
* ''Matthew''
** Miniprepped, digested, and ran on gels candidates for mCherry and EYFP with Kozak
** Several candidates possible for sequencing for EYFP and 3 candidates for mCherry
[[Image: Polasko_EYFPigem.jpg|300px|thumb|left]]
EYFP Results - although it's hard to gel w/o mlc. marker. The bands are the right size around 700bp and iGEM pSB1c3 backbone at 2kb
From Left: Lane 1 - 100 bp ladder. Did not show
Lane 2 - Uncut EYFP candidate 1
Lane 3 - EYFP candidate 1
Lane 4 - Uncut EYFP candidate 2
Lane 5 - EYFP candidate 2
Lane 6 - Uncut EYFP candidate 3
Lane 7 - EYFP candidate 3
Lane 8 - Uncut EYFP candidate 4
Lane 9 - EYFP candidate 4
[[Image: Polasko_mCherryigem.jpg|300px|thumb|left]]
mCherry Results - although it's hard to gel w/o mlc. marker. The band in Lane 5 the right size around 700bp with the iGEM pSB1C3 backbone at 2kb
From Left: Lane 1 - 100 bp ladder. Did not show
Lane 2 - Uncut mCherry candidate 1
Lane 3 - mCherry candidate 1
Lane 4 - Uncut mCherry candidate 2
Lane 5 - mCherry candidate 2
Lane 6 - Uncut mCherry candidate 3
Lane 7 - mCherry candidate 3
Lane 8 - Uncut mCherry candidate 4
Lane 9 - mCherry candidate 4
'''Week of September 19-25:''' * ''Elaine'' ** 09/17/10 Submitted GFP with Kozak sequence (PSB1C3) into registry ** 09/20/10 Mailed GFP with Kozak sequence (PSB1C3) to MIT. ** Streaked single colonies [RD29A + RFP (PSB1C3)] and did Cell cultures ** Miniprepped [RD29A + RFP (PSB1C3)]and digested with EcoR I and Pst I ** Ran uncut and digested [RD29A + RFP (PSB1C3)] into 1.2 Agarose Gel ** Submitted [RD29A + RFP (PSB1C3)] for sequencing * ''Randy and Vadim'' ** Concentrated gel purification sample from Sept. 16 ** Ligated ccdB gene into pSB1C3 vector ** Transformed ccdB+pSB1C3 ligation into DB3 cells ** Miniprepped ccdB+pSB1C3 ligation * Samantha ** PCR RD29A from pMA plasmid ** Confirm PCR reaction via agarose gel * ''Matthew'' ** Miniprepped and submitted candidates for sequencing for mCherry and EYFP with Kozak ** Sequences came back positive for EYFP and mCherry *** Point mutation in the mCherry but shouldn't affect the translation '''Week of September 26-October 2:''' * ''Elaine'' ** Miniprepped some more [RD29A + RFP (PSB1C3)] samples for submission * ''Randy and Vadim'' ** Digested and gel-analyzed ccdB-pSB1C3 ligation with EcoRI and Pst *** Digest yielded successful ligation ** Made Kan and Amp resistant plates ** Made additional LB broth ** Autoclaved labware and equipment ** Miniprepped ligations from Sept. 23 ** Submitted ligation for sequencing at the Nevada Genomics Center ** Made glycerol stock of ligations from Sept. 23 ** Transformed ligations from Sept. 23 into NEB cells *** Plates yielded no growth-ccdB ligation successful * Samantha ** Gel purification of RD29A PCR product * ''Matthew'' ** Submitted Kozak-mCherry and Kozak-EYFP to iGEM ** Cultured and miniprepped some of Elaine's GFP iGEM vector ** Miniprepped, digested, ethanol precipitated, and began ligation on GFP iGEM with 35S in Topo '''Week of October 3-9:''' * ''Elaine'' ** Topocloned PCR product of RD29A ** Streaked single colonies of RD29A (Topo Vector) ** Cell cultured single colonies of RD29A (Topo Vector) * ''Matthew'' ** Completed ligation and began transformation for 35S-GFP candidates ** Screened 35S-GFP candidates on chloramphenicol and kanamycin ** Cultured,miniprepped,digested, and ran on gel 20 candidates *** One candidate possible for sequencing [[Image: Polasko_35sgfp.jpg|300px|thumb|left]]
35S-GFP Composite Results - With careful inspection of my gels, I believed Lane 3 was a good candidate. I was looking for 2 bands, the composite at about 1.8kb and the pSB1C3 backbone at 2kb. Although, the image is not clean, I believed I saw a second band emerging in between the molecular markers which were 2kb and 1.6kb, making that emerging band at 1.8kb. Sequencing would prove me right
From Left: Lane 1 - 1 kb ladder.
Lane 2 - Uncut 35S-GFP candidate 1
Lane 3 - 35S-GFP candidate 1
Lane 4 - Uncut 35S-GFP candidate 2
Lane 5 - 35S-GFP candidate 2
Lane 6 - Uncut 35S-GFP candidate 3
Lane 7 - 35S-GFP candidate 3
Lane 8 - Uncut 35S-GFP candidate 4
Lane 9 - 35S-GFP candidate 4
Lane 10 - 35S-GFP candidate 5
** Submitted for sequencing 1 35S-GFP candidate '''Week of October 10-16:''' * ''Matthew'' ** Sequencing results came back positive for 35S-GFP ** Miniprepped 35S-GFP and made glycerol stock '''Week of October 17-23:''' * ''Matthew'' ** Submitted 35S-GFP to iGEM '''Week of October 24-30:'''
 "
"