Team:LMU-Munich/Cut'N'survive/Functional Principle
From 2010.igem.org
(Difference between revisions)
(→Case b) cells incorporated with construct 2) |
(→Construct 1) |
||
| Line 10: | Line 10: | ||
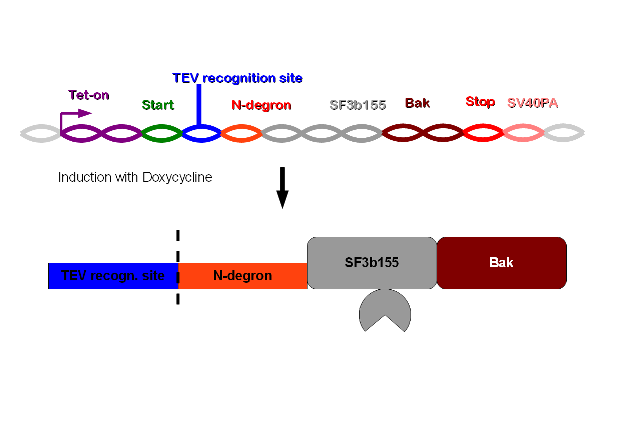
Tetracyclin-inducible promoter, TEV-recognition site, N-degron, SF3b155, human bak and SV40 Polyadenylation site. | Tetracyclin-inducible promoter, TEV-recognition site, N-degron, SF3b155, human bak and SV40 Polyadenylation site. | ||
| - | [[Image: | + | [[Image:cutwik32.jpg|600pxs|TEVdegron-System: TEV-degron-System]] |
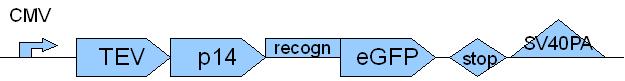
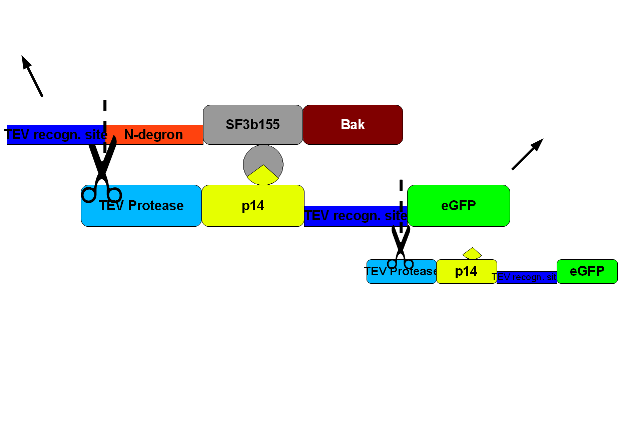
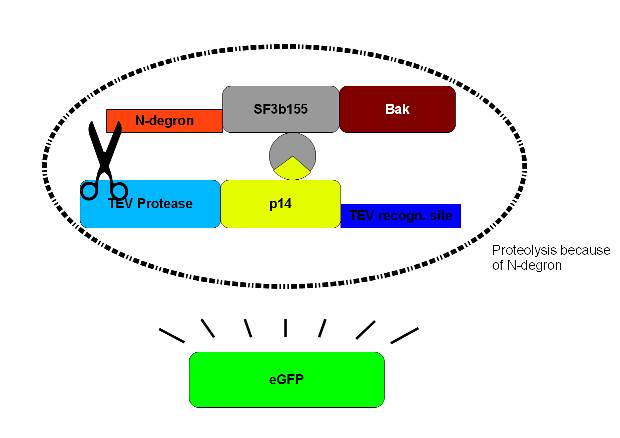
To select cells with this construct integrated into the cellular genome we perform a co-transfection with hygromycine resistence plasmid. The TEV-recognition site will be cut by the TEV-protease which is part of construct 2. Due to the digestion at the TEV-recognition site, the N-terminus of N-degron is free and thus provides a signal for the degeneration of the protein. SF3b155 is a protein interacting with p14* from construct 2. This interaction ascertains that the TEV-protease of construct 2 is being brought to the recognition site. The human bak is an efficient apoptosis inducing membrane protein. | To select cells with this construct integrated into the cellular genome we perform a co-transfection with hygromycine resistence plasmid. The TEV-recognition site will be cut by the TEV-protease which is part of construct 2. Due to the digestion at the TEV-recognition site, the N-terminus of N-degron is free and thus provides a signal for the degeneration of the protein. SF3b155 is a protein interacting with p14* from construct 2. This interaction ascertains that the TEV-protease of construct 2 is being brought to the recognition site. The human bak is an efficient apoptosis inducing membrane protein. | ||
Revision as of 16:28, 24 October 2010


![]()
![]()







![]()
 "
"