Team:Stockholm/18 October 2010
From 2010.igem.org
(→Andreas) |
NinaSchiller (Talk | contribs) (→PCR verification for Uppsala-Sweden team) |
||
| Line 85: | Line 85: | ||
PCR run ON. | PCR run ON. | ||
| + | |||
| + | ==Nina== | ||
| + | |||
| + | ===Polyacrylamide gel on SOD=== | ||
| + | |||
| + | I ran a polyacrylamide gel on SOD that I have tried to get pure. | ||
| + | |||
| + | Ladder: PageRuler Unst. Protein Ladder Fermentas | ||
| + | |||
| + | [[Image:Mlös1.jpg|200px]] | ||
| + | |||
| + | Arragement on gel: | ||
| + | |||
| + | [[Image:Aq6.jpg]] | ||
| + | |||
| + | I had a Coomassie blue staning and destaining on the gel. | ||
| + | |||
| + | [[Image:Aq5.jpg|250px]] | ||
| + | |||
| + | ===Protein concentration=== | ||
| + | |||
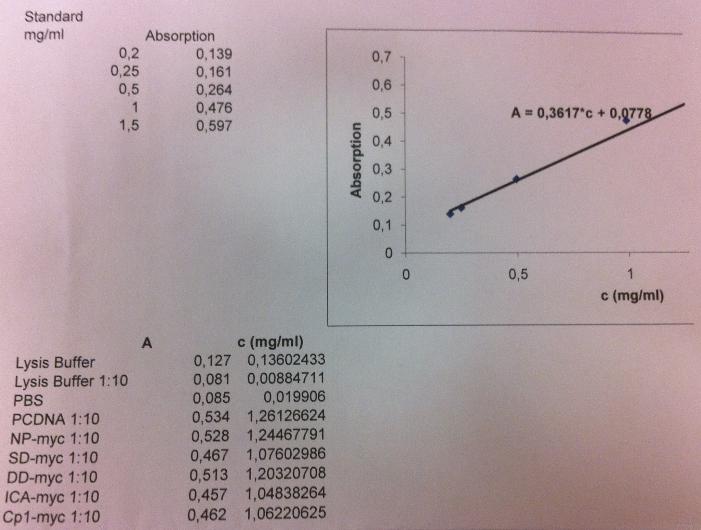
| + | I measured the protein conc on the samples: unbound, wash 1-3, elute 1-4 and not pure to check if I had any proteins. I had to test this since the gel shows that I don't have any proteins, even not much in the unbound lane, which should have a lot of bands. It could also be that my samples that I loaded onto the gel was very diluted since I had fractions consisting of 10 ml solution. This could give me an indication wheter I need to concentrate my fractions with a Millipore centrifugation. | ||
| + | |||
| + | I measured the concentrations using a BSA-method. | ||
| + | |||
| + | I got a standard curve from another student (Annika) in the lab I work in. | ||
| + | |||
| + | [[Image:Aq7.jpg|400px]] | ||
Revision as of 16:23, 24 October 2010
Contents |
Andreas
Transfer of ProtA⋅His to pEX
Digestion
| Sample | |
|---|---|
| 10X FastDigest buffer | 1 |
| Plasmid DNA | 7 |
| dH2O | 0 |
| FD XbaI | 1 |
| FD PstI | 1 |
| 10 μl |
- Incubation: 37 °C, 0:30
- Inactivation: 80 °C, 3:00
De-phosphorylated pre-digested/extracted pEX vector:
- 8 μl extracted and digested (X+P) pEX vector DNA
- 1 μl FD buffer
- 1 μl FastAP (alkaline phosphatase)
- Incubation: 37 °C, 0:30
- Inactivation: 80 °C, 3:00
Ligation
| 10X T4 Ligase buffer | 2 |
| Vector DNA | 1 |
| Insert DNA | 11 |
| dH2O | 5 |
| T4 DNA ligase | 1 |
| 20 μl |
|---|
- Incubation: 22 °C, 1 h
Transformation
Including three more transformations
Modified quick-transformation protocol:
- 30 min on ice
- 50 μl BL21
- 2 μl pEX.ProtA⋅his ligation mix
- 0.5 μl pEX.IgGp
- 0.5 μl pEX.SOD.RyC
- 0.5 μl pEX.hS.RyC
PCR verification for Uppsala-Sweden team
Helping the Uppsala team with a PCR verification of one of their assemblies.
Summed up their total construct length in pSB1x3 to 6566 bp.
- K1: C2 & C4 (x2)
- K2: C2 & C5 (x2)
Standard colony PCR settings:
- Elongation time: 10 min
- Annealing temp: 55 °C and 60 °C
PCR run ON.
Nina
Polyacrylamide gel on SOD
I ran a polyacrylamide gel on SOD that I have tried to get pure.
Ladder: PageRuler Unst. Protein Ladder Fermentas
Arragement on gel:
I had a Coomassie blue staning and destaining on the gel.
Protein concentration
I measured the protein conc on the samples: unbound, wash 1-3, elute 1-4 and not pure to check if I had any proteins. I had to test this since the gel shows that I don't have any proteins, even not much in the unbound lane, which should have a lot of bands. It could also be that my samples that I loaded onto the gel was very diluted since I had fractions consisting of 10 ml solution. This could give me an indication wheter I need to concentrate my fractions with a Millipore centrifugation.
I measured the concentrations using a BSA-method.
I got a standard curve from another student (Annika) in the lab I work in.
 "
"