Team:Wisconsin-Madison/delivery
From 2010.igem.org
Msagstetter (Talk | contribs) (→Bile Sensitive) |
(→Inducible-Repressible) |
||
| Line 134: | Line 134: | ||
[[Image:UW_IR_LuxR_induction.png|500px]] | [[Image:UW_IR_LuxR_induction.png|500px]] | ||
| + | |||
| + | After leaving the stomach, the repressor cI is degraded at a faster rate as compared to LuxR due to an attached LVA tag. This causes an increasing cell lysis signal. | ||
'''4) In Small Intestine''' | '''4) In Small Intestine''' | ||
[[Image:UW_IR_LuxR_induction.png|500px]] | [[Image:UW_IR_LuxR_induction.png|500px]] | ||
| + | |||
| + | Eventually, the LuxR:cI ratio is sufficiently high to cause the cell lysis signal via the inducible-repressible promoter. | ||
<br> | <br> | ||
Revision as of 18:41, 23 October 2010
Abstract
We have designed a universal platform for polypeptide release within the small intestine of the human gut that eliminates the need for any manual induction. Our automated, model system releases beta-galactosidase, a functional homologue of human lactase, once it reaches the duodenum to help a lactose intolerant patient metabolize lactose. The chassis for this system is the common probiotic in yoghurt, Lactobacillus acidophilus. Once the Lactobacillus acidophilus has reached the duodenum, they will lyse by either by a timed inducible/repressible system, a bile-inducible system, or an encryption system.
Background
Genetic Disorders
Lactose Intolerance
Even though our system has the potential to be applied to many medical conditions, our project is focusing on the goal of aiding in lactose digestion in lactose intolerance in children and adults. Lactose intolerance is the condition where people loose the ability to produce Beta-galactosidase enzyme, or lactase, in their intestines to help breakdown the lactose sugar present in many dairy products. According to the University of Virginia Health System, about 30 to 50 million Americans have lactose intolerance. This condition has two different types of lactose deficiency. Primary lactose deficiency is developed over time after the child is about two years old. However, the symptoms do not show until late adolescence and/or adulthood (NIH). Primary lactose deficiency has been found to be genetically linked and commonly found in certain racial demographics. Secondary lactose deficiency is a result from injury to the small intestines from disease or chemotherapy. Symptoms of lactose deficiency include abdominal pain and bloating, diarrhea and nausea (NIH).
Physiology
Small intestines
Beta-galactosidase enzyme is produced by the cells in the lining of the small intestines. The acidity of the small intestines ranges from pH 5 to 6. For the beta-galactosidase, we used the lacZ gene, which is a homolog of human lactase.
Stomach
The stomach is the part of the body where we hope to employ the colonic acid genes to form a protective capsule to increase bacterial survival until it reaches the small intestines. The ph of the stomach is around 2. However, in the presence of yoghurt, the yoghurt causes in the acidity of the stomach to be around pH 2.7 to 4. According to a study on the effects of yoghurt in the stomach, the pH was greater than 2.7 for 3 hour during digestion (Martini et. al., 1987).
Current Treatments
Probiotics
Probiotics http://nccam.nih.gov/health/probiotics/
For our project, we have chosen Lactobacillus acidophilus as the chassi to harbor this system because it is one of the common bacteria present in the human intestines and is commonly added to yoghurt as a probiotic. Probiotics have been gaining popularity in the market and in research due to the possible benefits it provides to the host. Medically, probiotics is considered as a living organism that is similar to the normal flora of the human gut and provides benefits to the host.
Modules
Enzyme Production
Our model system will tackle lactose intolerance. Producing Human Lactase in our bacterial cells would be difficult so we have chosen an enzyme that acts as functional homologue of human lactase called Beta-Galactacidase (B-Gal).
Left Lactase, Right Beta-Galactacide
We have taken advantage of the wealth of device that can be found in the iGEM Registry of Standard Parts. In 2008, the Cal Tech team had created a series of devices containing the gene for Beta-Galactacidase production. The difference in each device is the activity of each of the constitutive promoters. Thanks to their work, we can fine tune the amount of Beta-Galactacidase produces within our cells.
[IMAGE: RESULTS SHOWING CONSITUIVE EXPRESSION OF SERIES]
It is important to remember that this is the replaceable construct of our project. Because 1) testing for B-Gal activity on Lactose is easy and 2) the gene B-Gal is easily obtained and very well characterized, we have chosen to create a Lactose Intolerance specific treatment.
The following must be done to tackle another genetic disorder;
- Find the polypeptide specific to improving the condition of a genetic disorder
- EXAMPLE
- Produce a construct that allows the cell to overproduce this polypeptide
- Add to iDIET system
Capsule Basics
Our goals is to have each cell be surrounded by a protective 'capsule' to allow them to safely travel through the harsh acidic environment of the stomach to arrive in the small intestine for their main purpose. A past iGEM team used Transcription Factor RcsB to stimulate a the production of Colonic Acid from the capsule synthesis pathway of E.coli. Colonic Acid is a polymer of {=====] and has been shown to increase cell survivability in acidic conditions. We investigated the pathway for capsular polysaccharides synthesis and found more transcription factors that could give a similar or even better results!
[IMAGE: STOMACH INTESTINE]
RcsA and RcsB are transcription factors that are know to be positive regulators of capsular polysaccharides synthesis. We placed RcsA, RcsB, and a combination of the two under a IPTG inducible promoter to test both quantity of colonic acid produced and cell survivability. These two transcription factors form a heterodimer that is know to activate around 19 genes related to colonic acid synthesis. RcsB is also know to form a homodimmer and positively regulate cell division. RcsA and RcsB belong to the multicomponent RcsF/RcsC/RcsD/RcsA-RcsB phosphorelay system.
More information on these transcription factors and their usage in Biology can be found here: http://biocyc.org/ECOLI/NEW-IMAGE?type=GENE&object=EG10820 Ecocyc-RcsA & http://biocyc.org/ECOLI/NEW-IMAGE?type=GENE&object=EG10821 Ecocyc-RcsB
We wanted to test what IPTG induction levels were appropriate to protect our cells from an acidic environment of pH=2 before placing our devices under constitute control with appropriate ribosome binding sites.
[IMAGE OF 3 DEVICES] [CELL SURVIVABILITY] [COLONIC ACID #}\]
Based on our experiments quantifying the amount of colonic acid produces and cell survivability after acid shock, we have chosen [----------] to use in our final device which will be under constitutive promotion
Our final system will utililize a probiotic stain called Lactobacillus Acidophiles that is native to the human gut. The pathways in this bacterium are different than that of E.coli. Our next stage of research will involve finding a pathway, gene, or transcription factor that upregulates capsule synthesis and protocols to engineer this strain.
(IMAGE OF FINAL DEVICE]
One caution we had to consider in up-regulating this pathway is biofilm formation. Cholonic Acid synthesis is one of the first steps of the biofilm formation pathway. To ensure a biofilm is not produced we have taken advantage of an inhibitory transcription factor YgiV that blocks the activation of further steps in this pathway. For now, we are content to test our constructs in an E.Coli chassis.
Lysis Expression
Why we need lysis.
pH Sensitive
pH sensitive expression of LuxR and cI-LVA proteins will be accomplished via the glutamate decarboxylase A promoter (gadAp). Native to the E. coli acid resistance "island", regulation of expression from this promoter is normally accomplished by binding or unbinding of the Fis, GadX, and Crp proteins. Although the mechanism and exact conditions which bring about expression from the promoter are not well understood, studies have pointed to a combination of stationary phase and low environmental pH causing induction (Castanie-Cornet MP et al, 2001; 2010).
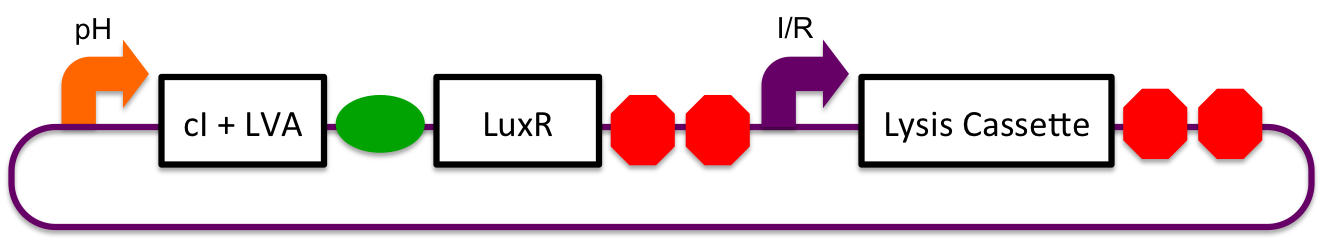
Inducible-Repressible
A previously characterized inducible-repressible promoter (BBa_R0065) will be used to accomplish our goal of delivering E. coli lactase to the human intestine. This system will be used in combination with the previously described gadA promoter, LuxR to yield expression after the active cell has seen the acidic conditions of the stomach. In order to better describe the order of events, refer to the following diagrams and descriptions:
1) Before Ingestion
The gadA promoter sees marginally acidic or neutral conditions and therefore yields expression of insignificant levels of both LuxR and cI-LVA. Since expression of both LuxR and cI-LVA are at low levels, the level of induction from inducible-repressible promoter is limited and cell lysis is not induced.
2) In Stomach
Upon arrival to the stomach, the acidic pH conditions experienced by the cells induces expression from the gadA promoter. LuxR and cI-LVA are both, therefore, expressed. In the presence of both the activator and repressor of the inducible-repressible promoter BBa_R0065, there is only very limited expression of the downstream genes. We believe that any "leaky" expression will be further reduced by use of a very weak activator (LuxR without the presence of molecules of AHL). A future step would be to remove the N-terminal end of the protein in order to mitigate potential complications due to accidental presence of AHL.
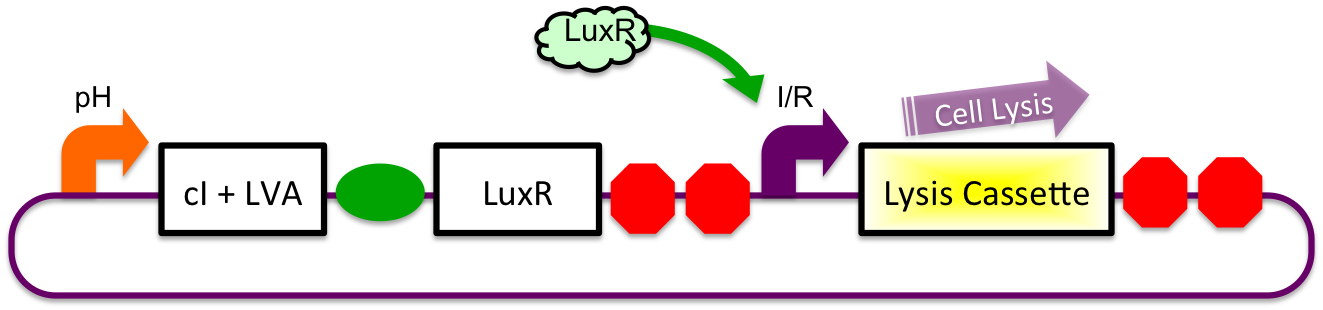
3) Immediately After Stomach
After leaving the stomach, the repressor cI is degraded at a faster rate as compared to LuxR due to an attached LVA tag. This causes an increasing cell lysis signal.
4) In Small Intestine
Eventually, the LuxR:cI ratio is sufficiently high to cause the cell lysis signal via the inducible-repressible promoter.
Bile Sensitive
Bile inducible expression
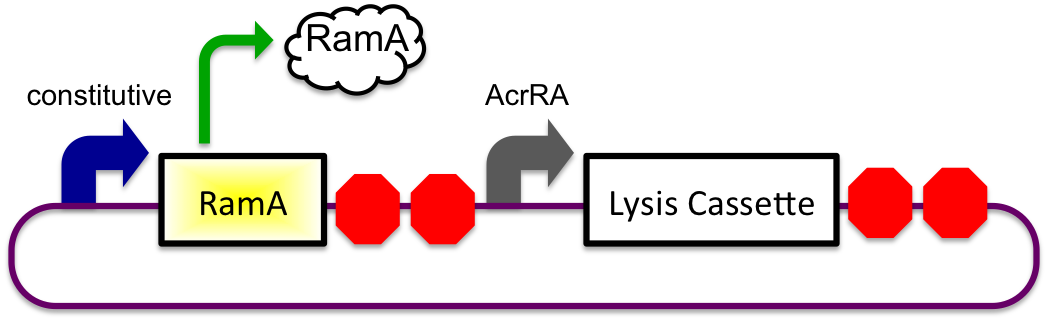
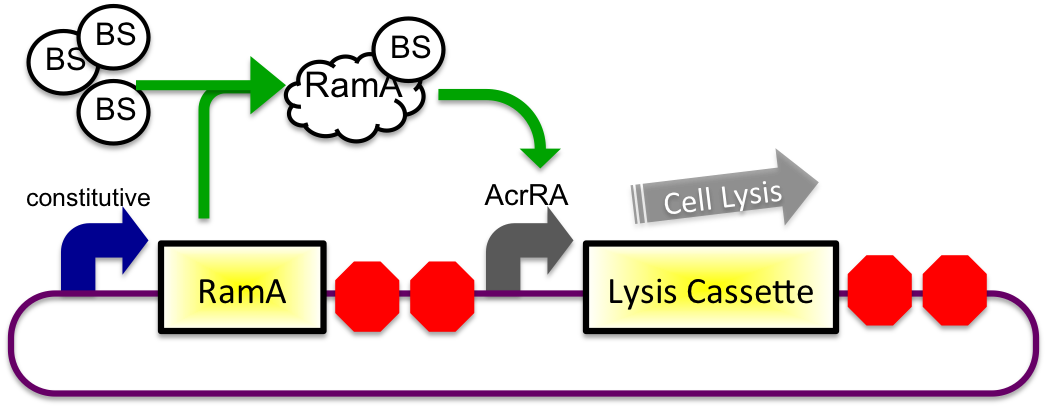
The bile inducible system consists of the ramA gene and the acrRA operon that were PCR amplified from Salmonella enterica strain LT2, locus tag STM0581. RamA is a regulatory protien that binds to bile and then binds to a specific sequence upstream of the acrAB operon(Nikaido et.al, 2008). This regulatory binding causes an increase in transcription of the acrAB gene (Nikaido et.al, 2008). To apply this concept to the timed lysis mechanism,the acrRA operon was cloned in front of a GFP reporter part, E0240, from the registry. Since E. coli DH10B cells do not endogenously produce ramA protein, I also cloned the ramA behind a constitutive promoter so the protein is constantly present in the cell to bind with bile. Ideally, these two constructs would then be cloned into plasmids with two different origins of replication for compatibility inside the cell.
In theory, when the cells come into contact with bile, the ramA will bind bile and then bind to the DNA, thus causing transcription of the acrA and the lysis gene, which will cause the cells to lyse and release beta-galactosidase. See images below.
Bioinformatics
Conserved domains http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi?RID=BKT8A21D014&mode=all Some of the conserved domains within the protein are HTHAraC and PRK 10219. HTHAraC is a bacterial regulatory helix-turn-helix protein. The Blast score was 78 and the location of the domain is 15-56 amino acids. The other conserved domain is PRK 10219, which is a DNA-binding transcriptional regulator SoxS. The blast score was 244 with the location to be 6-105 amino acids.
Homologues
From the Salmonella enterica LT2 sequence at NCBI, the RamA attachment site is located within the acrR gene. Based from Nikaido et.al., and from the Salmonella enterica LT2 sequence, the proposed attachment site is 5’-atggcac gaaaaa ccaaaca-3’. The Blast of the RamA protein sequence (NP_459573.1) resulted in similar proteins found in other Salmonella enterica species and subspecies, Citrobacter species, Klebsiella pneumonia, and Enterobacter species http://www.ncbi.nlm.nih.gov//blast/Blast.cgi . The blast of the ramA gene sequence (NC_003197.1) resulted in similar sequences found in Salmonella enterica subspecies enterica with different serovars, Citrobacter koseri ATCC BAA-895, and Citrobacter rodentium ICC168 (http://www.ncbi.nlm.nih.gov//blast/Blast.cgi).
1) Before Small Intestine
2) In Small Intestine
Enzyme Action
Complete System
Significance
Rare Genetic Disorders
Biopharmaceuticals
Protein purification
Enzyme Activity
Industry
Fermentation
Business Plan
Cost Effectiveness
Animation
 "
"