Team:Harvard/allergy/notebook
From 2010.igem.org
(Difference between revisions)
(→08-05-2010 [ top ]) |
|||
| Line 27: | Line 27: | ||
==06-14-2010 [ [[#top|top]] ]== | ==06-14-2010 [ [[#top|top]] ]== | ||
| - | * | + | * General Planning |
| - | * | + | * Discussion of RNAi |
| - | * | + | * Construct and order primers for sense and antisense parts |
| - | + | ||
| - | + | ||
'''Results/ Conclusions'''<br> | '''Results/ Conclusions'''<br> | ||
| - | + | ||
| - | + | Our goal is to work with Arabadopsis thaliana and strawberry DNA. We will extract gDNA and use PCR to obtain parts coding for allergens LTP, Bet, and Ger. | |
==06-15-2010 [ [[#top|top]] ]== | ==06-15-2010 [ [[#top|top]] ]== | ||
| + | * Extract gDNA of arabadopsis and strawberries | ||
| + | |||
| + | '''Results'''<br> | ||
| + | |||
| + | We were able to extract two samples of Arabadopsis gDNA (100 microliters of 1.5ng / microliter). We were not able to extract gDNA from strawberries. | ||
==06-16-2010 [ [[#top|top]] ]== | ==06-16-2010 [ [[#top|top]] ]== | ||
| + | * | ||
==06-17-2010 [ [[#top|top]] ]== | ==06-17-2010 [ [[#top|top]] ]== | ||
Revision as of 21:39, 22 October 2010

notebook calendar
06-14-2010 [ top ]
- General Planning
- Discussion of RNAi
- Construct and order primers for sense and antisense parts
Results/ Conclusions
Our goal is to work with Arabadopsis thaliana and strawberry DNA. We will extract gDNA and use PCR to obtain parts coding for allergens LTP, Bet, and Ger.
06-15-2010 [ top ]
- Extract gDNA of arabadopsis and strawberries
Results
We were able to extract two samples of Arabadopsis gDNA (100 microliters of 1.5ng / microliter). We were not able to extract gDNA from strawberries.
06-16-2010 [ top ]
06-17-2010 [ top ]
06-18-2010 [ top ]
06-21-2010 [ top ]
06-22-2010 [ top ]
06-23-2010 [ top ]
06-24-2010 [ top ]
06-25-2010 [ top ]
06-28-2010 [ top ]
06-29-2010 [ top ]
06-30-2010 [ top ]
07-01-2010 [ top ]
07-02-2010 [ top ]
07-05-2010 [ top ]
07-06-2010 [ top ]
07-07-2010 [ top ]
07-08-2010 [ top ]
07-09-2010 [ top ]
07-12-2010 [ top ]
07-13-2010 [ top ]
07-14-2010 [ top ]
07-15-2010 [ top ]
07-16-2010 [ top ]
07-19-2010 [ top ]
07-20-2010 [ top ]
07-21-2010 [ top ]
07-22-2010 [ top ]
07-23-2010 [ top ]
07-26-2010 [ top ]
07-27-2010 [ top ]
07-28-2010 [ top ]
07-29-2010 [ top ]
07-30-2010 [ top ]
08-02-2010 [ top ]
08-03-2010 [ top ]
- Grew up cultures of completed ihpRNA constructs (Bet, LTP, Ger) in pORE expression vector
- amiRNA PCR
- Will look at results of PCR tommorrow
08-04-2010 [ top ]
Tasks
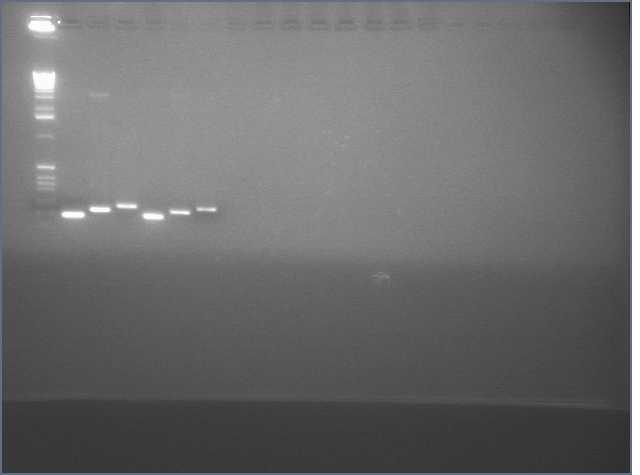
- amiRNA PCR appears to have worked at every Tm we tried:
- Digested V9/V10 to insert our constructs into
- Realized that we hadn't gel purified our ihpRNA inserts
- Gel purification of inserts (entire ihpRNA parts)
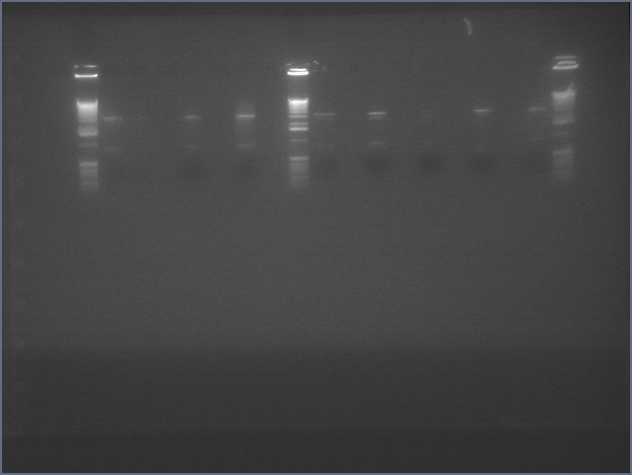
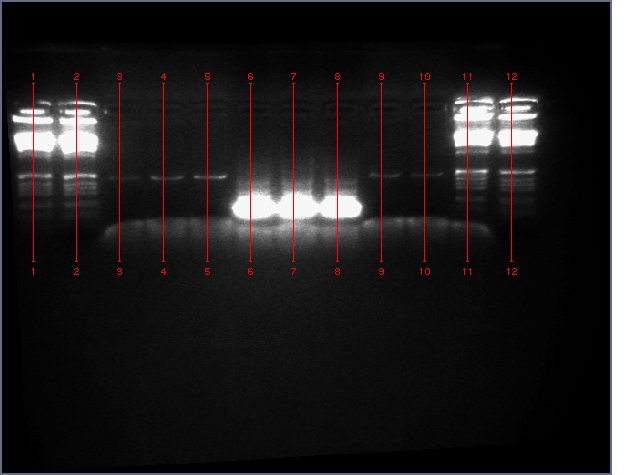
Ladder, 9, 11c1, 11c2, ladder, 25c1, 28c1, 28c2, 36c1, 36c2, ladder
Results
- Successfully gel purified our inserts and digested backbones that we will ligate into
08-05-2010 [ top ]
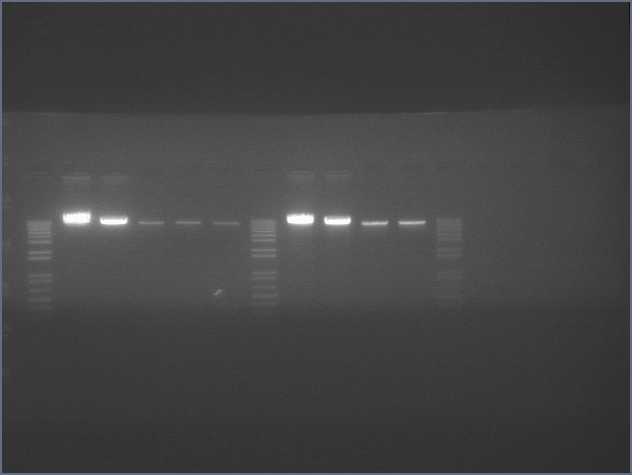
- Gel extracted V9/V10 backbone
Lanes: Ladder, V9, Ladder, V10
Concentrations: V9 (9.4 ng/uL; V10 (16.4 ng/uL)
- Ligated ihpRNA inserts into V9/V10 and transformed
- For our ligations we only used ~ 2uL of backbone (around 18 and 32 ng of backbone)and used a 3x excess of insert
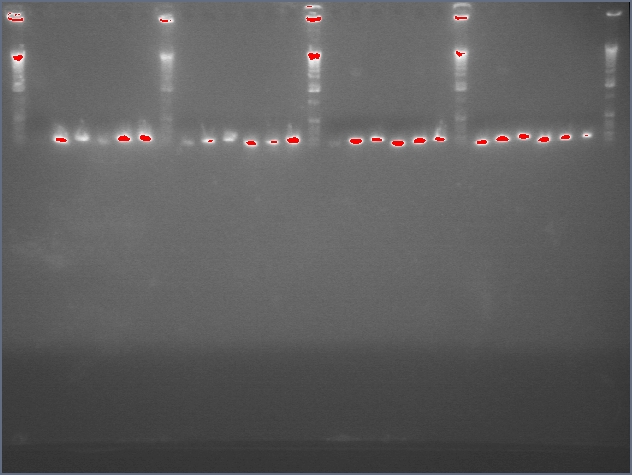
- Verified that amiRNA stitching of Bet, LTP yielded the proper insert with a low level of background through PCR:
Lanes: 1-7: Bet, corresponding to 65.55 degrees C for stitching Tm (even spacing)
Lanes: 8-10: LTP, corresponding to 65.55 degrees C during stitching annealing, even spacing
- Digested
- Bet,LTP inserts with X+P; B21 with X+P+phosphatase
- Ligated, Transformed
 "
"