Team:Stockholm/6 October 2010
From 2010.igem.org
NinaSchiller (Talk | contribs) (→Agarose gel) |
(→Andreas) |
||
| Line 10: | Line 10: | ||
*pSB1K3.nLMWP⋅SOD⋅His.RBS.yCCS 2 (VF2 & VR) | *pSB1K3.nLMWP⋅SOD⋅His.RBS.yCCS 2 (VF2 & VR) | ||
*pSB1K3.nLMWP⋅SOD⋅His.RBS.yCCS 3 (VF2 & VR) | *pSB1K3.nLMWP⋅SOD⋅His.RBS.yCCS 3 (VF2 & VR) | ||
| - | *pEX.nTra10⋅SOD⋅His (pEXf) | + | *pEX.nTra10⋅SOD⋅His (pEXf) ([[media:PEX.nT10SH_VF_premix_fasta_6oct.txt|fasta]]) |
| - | *pEX.nLMWP⋅SOD⋅His (pEXf) | + | *pEX.nLMWP⋅SOD⋅His (pEXf) ([[media:PEX.nLSH_VF_premix_fasta_6oct.txt|fasta]]) |
Sequences aligned using Geneious software, showing correct sequences for all constructs. Operon constructs all had an insertion between SpeI and PstI; this will not affect expression. | Sequences aligned using Geneious software, showing correct sequences for all constructs. Operon constructs all had an insertion between SpeI and PstI; this will not affect expression. | ||
| Line 66: | Line 66: | ||
---- | ---- | ||
| + | |||
==Nina== | ==Nina== | ||
Revision as of 16:15, 14 October 2010
Contents |
Andreas
Sequencing results
Received sequencing results for the following samples (fasta):
- pSB1K3.nTra10⋅SOD⋅His.RBS.yCCS 1 (VF2 & VR)
- pSB1K3.nTra10⋅SOD⋅His.RBS.yCCS 2 (VF2 & VR)
- pSB1K3.nTAT⋅SOD⋅His.RBS.yCCS 2 (VF2 & VR)
- pSB1K3.nTAT⋅SOD⋅His.RBS.yCCS 3 (VF2 & VR)
- pSB1K3.nLMWP⋅SOD⋅His.RBS.yCCS 2 (VF2 & VR)
- pSB1K3.nLMWP⋅SOD⋅His.RBS.yCCS 3 (VF2 & VR)
- pEX.nTra10⋅SOD⋅His (pEXf) (fasta)
- pEX.nLMWP⋅SOD⋅His (pEXf) (fasta)
Sequences aligned using Geneious software, showing correct sequences for all constructs. Operon constructs all had an insertion between SpeI and PstI; this will not affect expression.
Plasmid prep
Of stored pellets and ON cultures from 5/10
- E.Z.N.A. Plasmid Miniprep kit
- 50 μ elution volume (elution buffer)
| DNA concentration | ||
|---|---|---|
| Sample | Conc [ng/μl] | A260/A280 |
| pEX.nTAT⋅SOD⋅His.RBS.yCCS | 107.5 | 1.96 |
| pEX.nTra10⋅SOD⋅His.RBS.yCCS | 108.7 | 1.93 |
| pEX.nLMWP⋅SOD⋅His.RBS.yCCS | 84.35 | 1.97 |
| pSB1C3.His⋅SOD⋅cLMWP B | 255.4 | 1.94 |
| pSB1C3.His⋅SOD⋅cLMWP C | 201.2 | 1.94 |
| pSB1C3.His⋅SOD⋅cTAT D | 369.5 | 1.92 |
| pSB1C3.His⋅SOD⋅cTra10 A | 188.6 | 1.95 |
BL21 transformation
Quick transformation w/ 30 min on ice, 50 sec heat-shock.
- 30 μl competent BL21(DE3)
- 1 μl plasmid
- pEX.nTra10⋅SOD⋅His.RBS.yCCS
- pEX.nTAT⋅SOD⋅His.RBS.yCCS
- pEX.nLMWP⋅SOD⋅His.RBS.yCCS
Nina
Colony PCR on Fusion protein and protein A
I screened four colonies per dish.
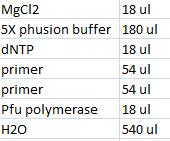
- Master mix with shipping vector verification primers (VF2 and VR):
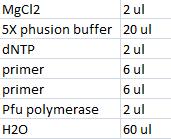
- Master mix with peX verification primers:
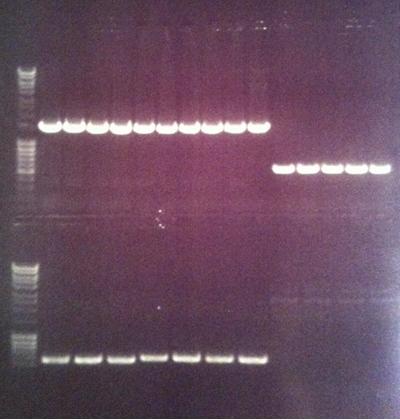
Agarose gel
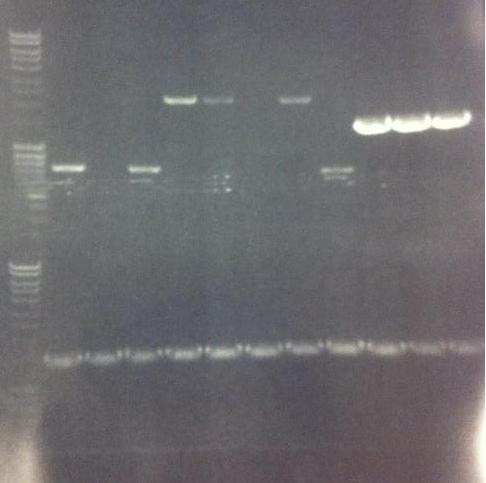
I ran the PCR products on an 1 % agarose gel (110 V) in order to confirm the inserts in the desired vectors.
Ladder: MassRuler™ DNA Ladder Mix, ready-to-use, 80-10,000 bp
Arrengement on the gels:
- Results of the gels:
All the Fusion and protein A bands look great, however the IgG protease bands look like they have not any insert which is bad. Tomorrow I'll run an agarose gel on my saved ligation sample of IgG protease in the gel cleaned peX vector. This will allow me to check for vector formation with a correct size and see if there has at least occured a ligation, if so I will run a new colony screen.
Mini prep on His-IgG protease and His-Protein A
I performed a mini prep on the inoculated colonies # 1, 2, 3 and 4 of N-terminal His-IgG protease and inoculated colonies # 1, 2, 3 and 4 of N-terminal His-Protein A.
 "
"