Team:Stockholm/7 October 2010
From 2010.igem.org
(→Removal of insertion in BioBrick suffixes) |
(→Andreas) |
||
| Line 12: | Line 12: | ||
*'''pEX.nLMWP⋅SH.Ry_pEXf:''' ASB0045 772 | *'''pEX.nLMWP⋅SH.Ry_pEXf:''' ASB0045 772 | ||
*'''pEX.nLMWP⋅SH.Ry_pEXf:''' ASB0045 773 | *'''pEX.nLMWP⋅SH.Ry_pEXf:''' ASB0045 773 | ||
| + | |||
| + | *15 μl plasmid DNA; 1.5 μl primer | ||
| + | |||
| + | {|border="1" cellpadding="1" cellspacing="0" | ||
| + | !colspan="3"|DNA concentration | ||
| + | |- | ||
| + | !Sample | ||
| + | !width="50"|Primer | ||
| + | !width="120"|Label | ||
| + | !width="120"|Sequence code | ||
| + | |- | ||
| + | |pEX.nTra10⋅SOD⋅His.RBS.yCCS | ||
| + | |align="center"|pEXf | ||
| + | |pEX.nTra10⋅SH.Ry_pEXf | ||
| + | |align="center"|ASB0045 768 | ||
| + | |- | ||
| + | |pEX.nTra10⋅SOD⋅His.RBS.yCCS | ||
| + | |align="center"|pEXr | ||
| + | |pEX.nTra10⋅SH.Ry_pEXr | ||
| + | |align="center"|ASB0045 769 | ||
| + | |- | ||
| + | |pEX.nTAT⋅SOD⋅His.RBS.yCCS | ||
| + | |align="center"|pEXf | ||
| + | |pEX.nTAT⋅SH.Ry_pEXf | ||
| + | |align="center"|ASB0045 770 | ||
| + | |- | ||
| + | |pEX.nTAT⋅SOD⋅His.RBS.yCCS | ||
| + | |align="center"|pEXr | ||
| + | |pEX.nTAT⋅SH.Ry_pEXr | ||
| + | |align="center"|ASB0045 771 | ||
| + | |- | ||
| + | |pEX.nLMWP⋅SOD⋅His.RBS.yCCS | ||
| + | |align="center"|pEXf | ||
| + | |pEX.nLMWP⋅SH.Ry_pEXf | ||
| + | |align="center"|ASB0045 772 | ||
| + | |- | ||
| + | |pEX.nLMWP⋅SOD⋅His.RBS.yCCS | ||
| + | |align="center"|pEXr | ||
| + | |pEX.nLMWP⋅SH.Ry_pEXr | ||
| + | |align="center"|ASB0045 773 | ||
| + | |} | ||
===Removal of insertion in BioBrick suffixes=== | ===Removal of insertion in BioBrick suffixes=== | ||
Revision as of 15:59, 14 October 2010
Contents |
Andreas
Transfer of nCPP⋅SOD⋅His.RBS.yCCS operons to pEX
Sequencing
15 μl plasmid DNA, 1.5 μl primer
- pEX.nTra10⋅SH.Ry_pEXf: ASB0045 768
- pEX.nTra10⋅SH.Ry_pEXr: ASB0045 769
- pEX.nTAT⋅SH.Ry_pEXf: ASB0045 770
- pEX.nTAT⋅SH.Ry_pEXf: ASB0045 771
- pEX.nLMWP⋅SH.Ry_pEXf: ASB0045 772
- pEX.nLMWP⋅SH.Ry_pEXf: ASB0045 773
- 15 μl plasmid DNA; 1.5 μl primer
| DNA concentration | |||
|---|---|---|---|
| Sample | Primer | Label | Sequence code |
| pEX.nTra10⋅SOD⋅His.RBS.yCCS | pEXf | pEX.nTra10⋅SH.Ry_pEXf | ASB0045 768 |
| pEX.nTra10⋅SOD⋅His.RBS.yCCS | pEXr | pEX.nTra10⋅SH.Ry_pEXr | ASB0045 769 |
| pEX.nTAT⋅SOD⋅His.RBS.yCCS | pEXf | pEX.nTAT⋅SH.Ry_pEXf | ASB0045 770 |
| pEX.nTAT⋅SOD⋅His.RBS.yCCS | pEXr | pEX.nTAT⋅SH.Ry_pEXr | ASB0045 771 |
| pEX.nLMWP⋅SOD⋅His.RBS.yCCS | pEXf | pEX.nLMWP⋅SH.Ry_pEXf | ASB0045 772 |
| pEX.nLMWP⋅SOD⋅His.RBS.yCCS | pEXr | pEX.nLMWP⋅SH.Ry_pEXr | ASB0045 773 |
Removal of insertion in BioBrick suffixes
An insertion between SpeI and PstI present in IgGp, bFGF, ProtA, yCCS and SOD needs to be removed before submission to the Registry. This will be done by digestion with SpeI (inside insertion) and moving digested gene into a new vector.
Digestions
No sample of SOD available for the moment, so this will be digested after plasmid prep.
| pC.IgGp (A) | pC.bFGF (B) | pC.ProtA (C) | pC.yCCS (D) | pC.RFP (E) | |
|---|---|---|---|---|---|
| 10X FastDigest buffer | 2 | 2 | 2 | 2 | 2 |
| DNA | 6 | 10 | 16 | 14 | 11 |
| dH2O | 10 | 5 | 0 | 2 | 5 |
| FD SpeI | 1 | 1 | 1 | 1 | 1 |
| FD EcoRI | 1 | 1 | 1 | 1 | 1 |
| 20 μl | 20 μl | 20 μl | 20 μl | 20 μl |
- Incubation: 37 °C, 30 min
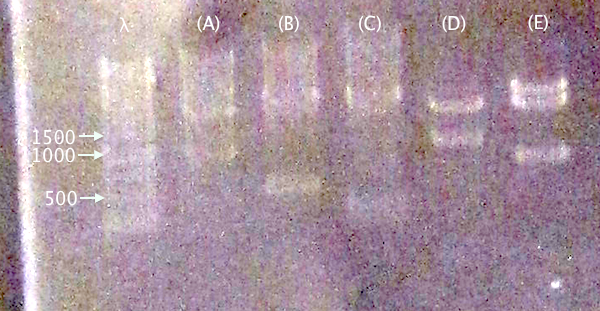
Gel verification
1 % agarose, 130 V
IgGp bFGF ProtA yCCS RFP
Expected bands
- pSB1C3.IgGp (A)
- Vector: 2050 bp
- Insert: 990 bp
- pSB1C3.bFGF (B)
- Vector: 2050 bp
- Insert: 520 bp
- pSB1C3.ProtA (C)
- Vector: 2050 bp
- Insert: 230 bp
- pSB1C3.yCCS (D)
- Vector: 2050 bp
- Insert: 800 bp
- pSB1C3.RFP (E)
- Vector: 2050 bp
- Insert: 1120 bp
Results
Relevant-sized for all samples showing successful (but incomplete) digestion. Proceeded to gel extraction.
Gel extraction
Loaded remaining 17 μl of each sample on a new 1 % agarose gel. Relevant bands excised by gel extraction and saved in -20 ° for later purification.
ON culture
Set ON culture of pSB1C3.SOD for later plasmid prep.
- 5 ml LB + Cm 25; 37 °C, 225 rpm
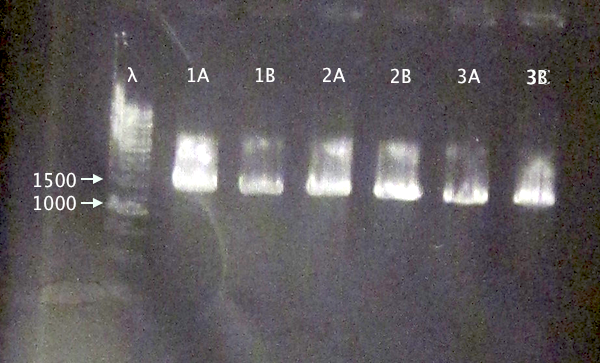
Colony PCR of nCPP⋅SOD⋅His.RBS.yCCS operons in BL21
- BL21 pEX.nTra10⋅SOD⋅His.RBS.yCCS (A & B)
- BL21 pEX.nTAT⋅SOD⋅His.RBS.yCCS (A & B)
- BL21 pEX.nLMWP⋅SOD⋅His.RBS.yCCS (A & B)
- Standard colony PCR settings.
- Elongation: 1:30 (too short?)
Gel verification
1 % agarose, 130 V
Expected bands
- 1553 bp
- 1523 bp
- 1532 bp
Results
Bands with relevant sizes for all clones.
ON cultures
3 ml LB + Amp 100
- B
- B
- B
Growth curve assay
A growth curve assay will be run to test the toxicity of our CPPs.
ON cultures
5 ml LB + Amp 100; 37 °C, 225 rpm
- BL21, pEX.SOD⋅His
- BL21, pEX.nTra10⋅SOD⋅His
- BL21, pEX.nTAT⋅SOD⋅His
- BL21, pEX.nLMWP⋅SOD⋅His
 "
"