Team:Kyoto/Notebook
From 2010.igem.org
(→4. Culture of 1-6-G, 1-12-O, and 1-23-L=) |
|||
| Line 2: | Line 2: | ||
==Index== | ==Index== | ||
==Notebook== | ==Notebook== | ||
| + | |||
<div class="note"> | <div class="note"> | ||
| + | |||
===Tuesday, July 20=== | ===Tuesday, July 20=== | ||
| + | |||
By: Wataru, Tomo, Yuki, Kazuya, Ken, Makoto | By: Wataru, Tomo, Yuki, Kazuya, Ken, Makoto | ||
| + | |||
====1. Solubilization of antibiotics.==== | ====1. Solubilization of antibiotics.==== | ||
| - | + | ||
| - | + | For Ampicillin(Amp): add 1.0g Amp to 20ml MilliQ (final concentration is 50mg/ml). | |
| - | + | ||
| - | + | For Kanamycin(kan): add 0.5g Kan to 10ml MilliQ (final concentration is 50mg/ml). | |
| + | |||
| + | Dispense 1.1ml of the solution into 1.5ml tubes. | ||
| + | |||
| + | Store in the freezer (-20℃). | ||
| + | |||
====2. Making plates for LB (Amp+) and LB (Kan+).==== | ====2. Making plates for LB (Amp+) and LB (Kan+).==== | ||
| + | |||
====3. [[Team:Kyoto/Protocols#Transformation|Transformation]] of iGEM Parts.==== | ====3. [[Team:Kyoto/Protocols#Transformation|Transformation]] of iGEM Parts.==== | ||
| + | |||
{| class="experiments" | {| class="experiments" | ||
!Name||Well||Sample (µl)||Competent Cells (µl)||Total (µl)||Plate||Incubation||Result | !Name||Well||Sample (µl)||Competent Cells (µl)||Total (µl)||Plate||Incubation||Result | ||
| Line 31: | Line 42: | ||
|<partinfo>B0015</partinfo>||1-23-L||1||20||21||LB (Kanamycin+)||× | |<partinfo>B0015</partinfo>||1-23-L||1||20||21||LB (Kanamycin+)||× | ||
|} | |} | ||
| - | + | ||
| - | + | A vector of "pSB4K5" is Kanamycin-resistance, however, we plated it to LB plate (Ampicillin+). And We didn't pre-culture "B0015" despite its vector is Kanamycin-resistance. So, it was predicted that we will fail the transformation of "pSB4K5" and "B0015". | |
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
<div class="note"> | <div class="note"> | ||
===Wednesday, July 21=== | ===Wednesday, July 21=== | ||
| + | |||
By: Wataru, Ken, Makoto, Takuya Yamamoto | By: Wataru, Ken, Makoto, Takuya Yamamoto | ||
| + | |||
====1. Culture plates in which colonies was observed at 37℃ from 07/21 20:50 to 07/22 17:00.==== | ====1. Culture plates in which colonies was observed at 37℃ from 07/21 20:50 to 07/22 17:00.==== | ||
| + | |||
====2. Make a master plate of the above plates.==== | ====2. Make a master plate of the above plates.==== | ||
| + | |||
====3. Retry [[Team:Kyoto/Protocols#Transformation|Transformation]] of iGEM Parts.==== | ====3. Retry [[Team:Kyoto/Protocols#Transformation|Transformation]] of iGEM Parts.==== | ||
| + | |||
{| class="experiments" | {| class="experiments" | ||
!Name||Well||Sample (µl)||Competent Cells (µl)||Total (µl)||Plate||Incubation||Result | !Name||Well||Sample (µl)||Competent Cells (µl)||Total (µl)||Plate||Incubation||Result | ||
| Line 51: | Line 65: | ||
|<partinfo>B0015</partinfo>||1-23-L||1||20||21||○ | |<partinfo>B0015</partinfo>||1-23-L||1||20||21||○ | ||
|} | |} | ||
| + | |||
====4. [[Team:Kyoto/Protocols#PCR|PCR]] for S-R-Rz/Rz1 and S==== | ====4. [[Team:Kyoto/Protocols#PCR|PCR]] for S-R-Rz/Rz1 and S==== | ||
| - | + | ||
| + | Dilute λDNA (0.5µg/µl) 100 times with MilliQ. The final concentration of template λDNA was 5ng/µl. | ||
| + | |||
{| class="experiments" | {| class="experiments" | ||
!No.||Water||25mM MgSO4||2mM dNTPs||10xBuffer for KOD Plus ver.2||TemplateDNA (5ng/µl)||Primer S-R-Rz/Rz1 Forward (10µM)||Primer S-R-Rz/Rz1 Reverse (10µM)||Primer S Reverse (10µM)||KOD Plus ver.2||Total | !No.||Water||25mM MgSO4||2mM dNTPs||10xBuffer for KOD Plus ver.2||TemplateDNA (5ng/µl)||Primer S-R-Rz/Rz1 Forward (10µM)||Primer S-R-Rz/Rz1 Reverse (10µM)||Primer S Reverse (10µM)||KOD Plus ver.2||Total | ||
| Line 72: | Line 89: | ||
|8||28µl||3µl||5µl||5µl||5µl||1.5µl||-||1.5µl||1µl||50µl | |8||28µl||3µl||5µl||5µl||5µl||1.5µl||-||1.5µl||1µl||50µl | ||
|} | |} | ||
| - | + | ||
| - | + | Forward Primer of S-R-Rz/Rz1 and S is common. | |
| + | |||
| + | PCR condition : 94℃ x 2min, (98℃ x 10sec, 55℃ x 30sec, 68℃ x 1min) x 30cycles, 4℃ forever. | ||
| + | |||
</div> | </div> | ||
<div class="note"> | <div class="note"> | ||
| + | |||
===Thursday, July 22=== | ===Thursday, July 22=== | ||
| + | |||
By: Wataru | By: Wataru | ||
| + | |||
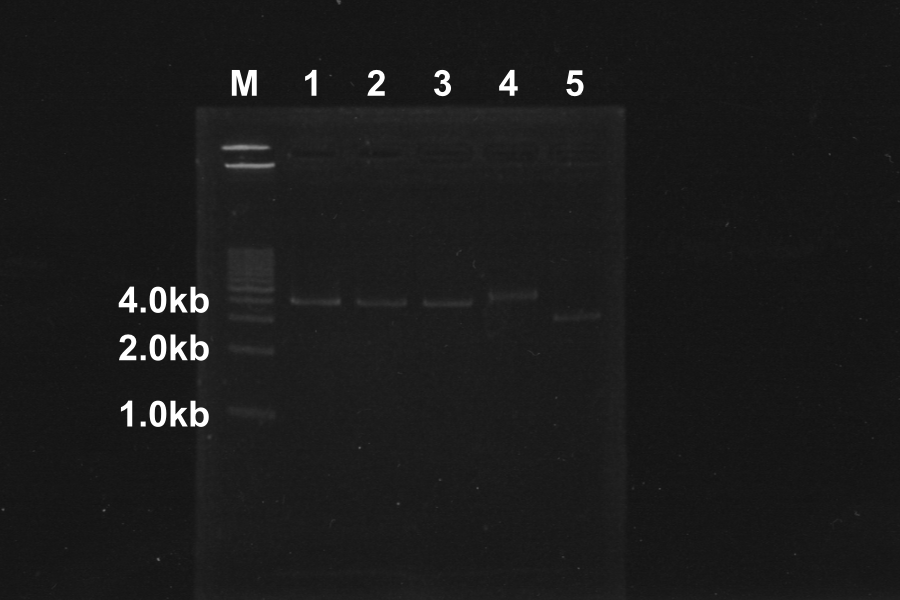
====1. [[Team:Kyoto/Protocols#Electrophoresis|Electrophoresis]] of the PCR products for 40min.==== | ====1. [[Team:Kyoto/Protocols#Electrophoresis|Electrophoresis]] of the PCR products for 40min.==== | ||
| + | |||
[[Image:KyotoEXP100722-1.png|right]] | [[Image:KyotoEXP100722-1.png|right]] | ||
| - | + | ||
| - | + | Length of S and S-R-Rz/Rz1 is 370bp, 1300bp, so PCR succeeded. | |
| + | |||
====2. [[Team:Kyoto/Protocols#Miniprep|Miniprep]] of iGEM Parts.==== | ====2. [[Team:Kyoto/Protocols#Miniprep|Miniprep]] of iGEM Parts.==== | ||
| + | |||
{| class="experiments" | {| class="experiments" | ||
!Name||Concentration(ng/µl) | !Name||Concentration(ng/µl) | ||
| Line 99: | Line 125: | ||
|<partinfo>J06702</partinfo>||14.7 | |<partinfo>J06702</partinfo>||14.7 | ||
|} | |} | ||
| - | + | ||
| - | + | The concentration of all samples was very week. Probably our shaking incubation was week. | |
| + | |||
====3. Culture plates and make master plates of <partinfo>pSB4K5</partinfo> and <partinfo>B0015</partinfo> from 7/22 17:00 to 7/23 10:00.==== | ====3. Culture plates and make master plates of <partinfo>pSB4K5</partinfo> and <partinfo>B0015</partinfo> from 7/22 17:00 to 7/23 10:00.==== | ||
| + | |||
</div> | </div> | ||
<div class="note"> | <div class="note"> | ||
| + | |||
===Friday, July 23=== | ===Friday, July 23=== | ||
| + | |||
By: Wataru, Tomo, Makoto | By: Wataru, Tomo, Makoto | ||
| + | |||
====1. [[Team:Kyoto/Protocols#Miniprep|Miniprep]] of iGEM Parts.==== | ====1. [[Team:Kyoto/Protocols#Miniprep|Miniprep]] of iGEM Parts.==== | ||
{| class="experiments" | {| class="experiments" | ||
| + | |||
!Name||Concentration(ng/µl) | !Name||Concentration(ng/µl) | ||
|- | |- | ||
| Line 115: | Line 147: | ||
|<partinfo>B0015</partinfo>||- | |<partinfo>B0015</partinfo>||- | ||
|} | |} | ||
| - | + | ||
| - | + | We lost <partinfo>B0015</partinfo> by our mistake. The concentration of <partinfo>pSB4K5</partinfo> is high, so this condition of shaking incubation is moderate. | |
| - | + | ||
====2. Picked up 1, 3, 5, 7 of the products of PCR, and purified by PCR-purification.==== | ====2. Picked up 1, 3, 5, 7 of the products of PCR, and purified by PCR-purification.==== | ||
| + | |||
{| class="experiments" | {| class="experiments" | ||
!Sample||Concentration (ng/µl)||New Name|| | !Sample||Concentration (ng/µl)||New Name|| | ||
| Line 130: | Line 163: | ||
|7||65.4||S<sub>2</sub> | |7||65.4||S<sub>2</sub> | ||
|} | |} | ||
| - | + | ||
| - | + | The concentration of sample number 1 and 5, the PCR products of S-R-Rz/Rz1, is week, so we desided to retry PCR. | |
| + | |||
====3. Retry of PCR of S-R-Rz/Rz1.==== | ====3. Retry of PCR of S-R-Rz/Rz1.==== | ||
| + | |||
{| class="experiments" | {| class="experiments" | ||
!Sample||Water||25mmol/l MgSO4||2mmol/l dNTPs||10×Buffer for KOD plus ver.2||Template DNA (5ng/µl)||Primer S-R-Rz/Rz1 Forward (10µmol/l)||Primer S-R-Rz/Rz1 Reverse (10µmol/l)||KOD plus ver.2||Total | !Sample||Water||25mmol/l MgSO4||2mmol/l dNTPs||10×Buffer for KOD plus ver.2||Template DNA (5ng/µl)||Primer S-R-Rz/Rz1 Forward (10µmol/l)||Primer S-R-Rz/Rz1 Reverse (10µmol/l)||KOD plus ver.2||Total | ||
| Line 148: | Line 183: | ||
|6||25||6||5||5||5||1.5||1.5||1||50 | |6||25||6||5||5||5||1.5||1.5||1||50 | ||
|} | |} | ||
| - | + | ||
| + | PCR condition : 94℃ x 2min, (98℃ x 10sec, 55℃ x 30sec, 68℃ x 1min) x 30cycles, 4℃ forever. | ||
| + | |||
====4. Digested <partinfo>J06702</partinfo> by EcoRI, XbaI, SpeI, PstI to check function of our Restriction enzymes.==== | ====4. Digested <partinfo>J06702</partinfo> by EcoRI, XbaI, SpeI, PstI to check function of our Restriction enzymes.==== | ||
| + | |||
{| class="experiments" | {| class="experiments" | ||
!Sample||10xBuffer||BSA||Enzyme||MilliQ||Total||Incubation | !Sample||10xBuffer||BSA||Enzyme||MilliQ||Total||Incubation | ||
| Line 165: | Line 203: | ||
====5. Electrophoresis of above sample for 35min.==== | ====5. Electrophoresis of above sample for 35min.==== | ||
| + | |||
[[Image:KyotoExp100723-1.png|right]] | [[Image:KyotoExp100723-1.png|right]] | ||
| - | + | ||
| - | + | Comparison to sample 5(control, circular DNA), the bands of sample 1, 2, 3, 4 was shifted. The DNA of sample 1, 2, 3, 4 was linearized by Restriction enzymes. So, our restriction enzymes work correctly. | |
| + | |||
====6. To insert S gene to GFP, we digested the PCR products of S gene by EcoRi and SpeI, and GFP by EcoRl and XbaI.==== | ====6. To insert S gene to GFP, we digested the PCR products of S gene by EcoRi and SpeI, and GFP by EcoRl and XbaI.==== | ||
| + | |||
{| class="experiments" | {| class="experiments" | ||
!Sample||10×Buffer||Enzyme 1||Enzyme 2||MilliQ||Total||Incubation | !Sample||10×Buffer||Enzyme 1||Enzyme 2||MilliQ||Total||Incubation | ||
| Line 178: | Line 219: | ||
|<partinfo>E0840</partinfo>(GFP)||45||5||''EcoR''I 0.2||''Xba''I 0.2||0||50 | |<partinfo>E0840</partinfo>(GFP)||45||5||''EcoR''I 0.2||''Xba''I 0.2||0||50 | ||
|} | |} | ||
| - | + | ||
| + | After PCR purification, evaporated them and diluted 3ul. | ||
| + | |||
====7. Ligated over night.==== | ====7. Ligated over night.==== | ||
| + | |||
{| class="experiments" | {| class="experiments" | ||
!Sample||Vector||Insert||Ligation High||Total | !Sample||Vector||Insert||Ligation High||Total | ||
| Line 187: | Line 231: | ||
|S-GFP<sub>2</sub>||<partinfo>E0840</partinfo> 0.5||S<sub>2</sub> 0.5||1||2 | |S-GFP<sub>2</sub>||<partinfo>E0840</partinfo> 0.5||S<sub>2</sub> 0.5||1||2 | ||
|} | |} | ||
| + | |||
</div> | </div> | ||
<div class="note"> | <div class="note"> | ||
| + | |||
===Monday, July 26=== | ===Monday, July 26=== | ||
| + | |||
By: Wataru, Tomonori, Makoto | By: Wataru, Tomonori, Makoto | ||
| + | |||
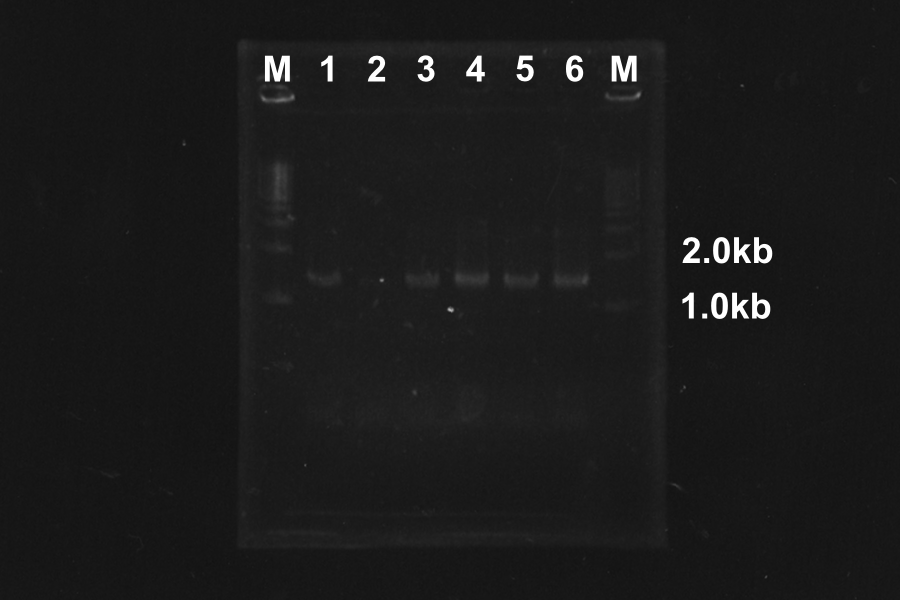
====1. Electrophoresis of PCR products==== | ====1. Electrophoresis of PCR products==== | ||
| + | |||
[[Image:KyotoExp100726-1.png|right]] | [[Image:KyotoExp100726-1.png|right]] | ||
| - | + | ||
| - | + | At the condition 4 (4.5µl MgSO4) and 6 (6µl MgSO4), S-R-Rz/Rz1 is amplified very much. So we decided to use them. | |
| + | |||
====2. PCR purification==== | ====2. PCR purification==== | ||
| + | |||
{| class="experiments" | {| class="experiments" | ||
!Sample||Concentration (ng/µl)||New Name | !Sample||Concentration (ng/µl)||New Name | ||
| Line 206: | Line 257: | ||
|6||59.6|| | |6||59.6|| | ||
|} | |} | ||
| + | |||
====3. Transformation of iGEM Parts==== | ====3. Transformation of iGEM Parts==== | ||
| + | |||
{| class="experiments" | {| class="experiments" | ||
!Name||Well||Sample (µl)||Competent Cell (µl)||Total (µl)||Plate||Incubation||Result | !Name||Well||Sample (µl)||Competent Cell (µl)||Total (µl)||Plate||Incubation||Result | ||
| Line 216: | Line 269: | ||
|||1-5-E||1||20||21||× | |||1-5-E||1||20||21||× | ||
|} | |} | ||
| + | |||
====4. Culture of 1-6-G, 1-12-O, and 1-23-L==== | ====4. Culture of 1-6-G, 1-12-O, and 1-23-L==== | ||
| + | |||
</div> | </div> | ||
---- | ---- | ||
Revision as of 09:51, 9 September 2010
Index
Notebook
Tuesday, July 20
By: Wataru, Tomo, Yuki, Kazuya, Ken, Makoto
1. Solubilization of antibiotics.
For Ampicillin(Amp): add 1.0g Amp to 20ml MilliQ (final concentration is 50mg/ml).
For Kanamycin(kan): add 0.5g Kan to 10ml MilliQ (final concentration is 50mg/ml).
Dispense 1.1ml of the solution into 1.5ml tubes.
Store in the freezer (-20℃).
2. Making plates for LB (Amp+) and LB (Kan+).
3. Transformation of iGEM Parts.
| Name | Well | Sample (µl) | Competent Cells (µl) | Total (µl) | Plate | Incubation | Result |
|---|---|---|---|---|---|---|---|
| <partinfo>J23100</partinfo> | 1-18-C | 1 | 20 | 21 | LB (Amp+) | At 37℃ 7/20 20:50 - 7/21 17:00 | ○ |
| <partinfo>J23105</partinfo> | 1-18-M | 1 | 20 | 21 | ○ | ||
| <partinfo>J23116</partinfo> | 1-20-M | 1 | 20 | 21 | ○ | ||
| <partinfo>R0011</partinfo> | 1-6-G | 1 | 20 | 21 | ○ | ||
| <partinfo>E0840</partinfo> | 1-12-O | 1 | 20 | 21 | ○ | ||
| <partinfo>J06702</partinfo> | 2-8-E | 1 | 20 | 21 | ○ | ||
| <partinfo>pSB4K5</partinfo> | 1-5-G | 1 | 20 | 21 | × | ||
| <partinfo>B0015</partinfo> | 1-23-L | 1 | 20 | 21 | LB (Kanamycin+) | × |
A vector of "pSB4K5" is Kanamycin-resistance, however, we plated it to LB plate (Ampicillin+). And We didn't pre-culture "B0015" despite its vector is Kanamycin-resistance. So, it was predicted that we will fail the transformation of "pSB4K5" and "B0015".
Wednesday, July 21
By: Wataru, Ken, Makoto, Takuya Yamamoto
1. Culture plates in which colonies was observed at 37℃ from 07/21 20:50 to 07/22 17:00.
2. Make a master plate of the above plates.
3. Retry Transformation of iGEM Parts.
| Name | Well | Sample (µl) | Competent Cells (µl) | Total (µl) | Plate | Incubation | Result |
|---|---|---|---|---|---|---|---|
| <partinfo>pSB4K5</partinfo> | 1-5-G | 1 | 20 | 21 | LB (Kanamycin+) | At 37℃ 7/21 20:50 - 7/22 16:30 | ○ |
| <partinfo>B0015</partinfo> | 1-23-L | 1 | 20 | 21 | ○ |
4. PCR for S-R-Rz/Rz1 and S
Dilute λDNA (0.5µg/µl) 100 times with MilliQ. The final concentration of template λDNA was 5ng/µl.
| No. | Water | 25mM MgSO4 | 2mM dNTPs | 10xBuffer for KOD Plus ver.2 | TemplateDNA (5ng/µl) | Primer S-R-Rz/Rz1 Forward (10µM) | Primer S-R-Rz/Rz1 Reverse (10µM) | Primer S Reverse (10µM) | KOD Plus ver.2 | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 28µl | 3µl | 5µl | 5µl | 5µl | 1.5µl | 1.5µl | - | 1µl | 50µl |
| 2 | 28µl | 3µl | 5µl | 5µl | 5µl | 1.5µl | 1.5µl | - | 1µl | 50µl |
| 3 | 28µl | 3µl | 5µl | 5µl | 5µl | 1.5µl | - | 1.5µl | 1µl | 50µl |
| 4 | 28µl | 3µl | 5µl | 5µl | 5µl | 1.5µl | - | 1.5µl | 1µl | 50µl |
| 5 | 28µl | 3µl | 5µl | 5µl | 5µl | 1.5µl | 1.5µl | - | 1µl | 50µl |
| 6 | 28µl | 3µl | 5µl | 5µl | 5µl | 1.5µl | 1.5µl | - | 1µl | 50µl |
| 7 | 28µl | 3µl | 5µl | 5µl | 5µl | 1.5µl | - | 1.5µl | 1µl | 50µl |
| 8 | 28µl | 3µl | 5µl | 5µl | 5µl | 1.5µl | - | 1.5µl | 1µl | 50µl |
Forward Primer of S-R-Rz/Rz1 and S is common.
PCR condition : 94℃ x 2min, (98℃ x 10sec, 55℃ x 30sec, 68℃ x 1min) x 30cycles, 4℃ forever.
Thursday, July 22
By: Wataru
1. Electrophoresis of the PCR products for 40min.
Length of S and S-R-Rz/Rz1 is 370bp, 1300bp, so PCR succeeded.
2. Miniprep of iGEM Parts.
| Name | Concentration(ng/µl) |
|---|---|
| <partinfo>J23100</partinfo> | 18.5 |
| <partinfo>J23105</partinfo> | 12.5 |
| <partinfo>J23116</partinfo> | 14.6 |
| <partinfo>R0011</partinfo> | 8.6 |
| <partinfo>E0840</partinfo> | 12.1 |
| <partinfo>J06702</partinfo> | 14.7 |
The concentration of all samples was very week. Probably our shaking incubation was week.
3. Culture plates and make master plates of <partinfo>pSB4K5</partinfo> and <partinfo>B0015</partinfo> from 7/22 17:00 to 7/23 10:00.
Friday, July 23
By: Wataru, Tomo, Makoto
1. Miniprep of iGEM Parts.
| Name | Concentration(ng/µl) |
|---|---|
| <partinfo>pSB4K5</partinfo> | 79.2 |
| <partinfo>B0015</partinfo> | - |
We lost <partinfo>B0015</partinfo> by our mistake. The concentration of <partinfo>pSB4K5</partinfo> is high, so this condition of shaking incubation is moderate.
2. Picked up 1, 3, 5, 7 of the products of PCR, and purified by PCR-purification.
| Sample | Concentration (ng/µl) | New Name | |
|---|---|---|---|
| 1 | 18.6 | - | |
| 3 | 77.6 | S1 | |
| 5 | 33.6 | - | |
| 7 | 65.4 | S2 |
The concentration of sample number 1 and 5, the PCR products of S-R-Rz/Rz1, is week, so we desided to retry PCR.
3. Retry of PCR of S-R-Rz/Rz1.
| Sample | Water | 25mmol/l MgSO4 | 2mmol/l dNTPs | 10×Buffer for KOD plus ver.2 | Template DNA (5ng/µl) | Primer S-R-Rz/Rz1 Forward (10µmol/l) | Primer S-R-Rz/Rz1 Reverse (10µmol/l) | KOD plus ver.2 | Total |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 28µl | 3 | 5 | 5 | 5 | 1.5 | 1.5 | 1 | 50 |
| 2 | 28 | 3 | 5 | 5 | 5 | 1.5 | 1.5 | 1 | 50 |
| 3 | 26.5 | 4.5 | 5 | 5 | 5 | 1.5 | 1.5 | 1 | 50 |
| 4 | 26.5 | 4.5 | 5 | 5 | 5 | 1.5 | 1.5 | 1 | 50 |
| 5 | 25 | 6 | 5 | 5 | 5 | 1.5 | 1.5 | 1 | 50 |
| 6 | 25 | 6 | 5 | 5 | 5 | 1.5 | 1.5 | 1 | 50 |
PCR condition : 94℃ x 2min, (98℃ x 10sec, 55℃ x 30sec, 68℃ x 1min) x 30cycles, 4℃ forever.
4. Digested <partinfo>J06702</partinfo> by EcoRI, XbaI, SpeI, PstI to check function of our Restriction enzymes.
| Sample | 10xBuffer | BSA | Enzyme | MilliQ | Total | Incubation |
|---|---|---|---|---|---|---|
| 1 | 5µl | 1 | EcoRI 0.1 | 3.6 | 10 | At 37℃ 7/23 18:00 - 7/23 18:30 |
| 2 | 5 | 1 | XbaI 0.1 | 3.6 | 10 | |
| 3 | 5 | 1 | SpeI 0.1 | 3.6 | 10 | |
| 4 | 5 | 1 | PstI 0.1 | 3.6 | 10 | |
| 5 | 5 | 1 | - | 3.7 | 10 |
5. Electrophoresis of above sample for 35min.
Comparison to sample 5(control, circular DNA), the bands of sample 1, 2, 3, 4 was shifted. The DNA of sample 1, 2, 3, 4 was linearized by Restriction enzymes. So, our restriction enzymes work correctly.
6. To insert S gene to GFP, we digested the PCR products of S gene by EcoRi and SpeI, and GFP by EcoRl and XbaI.
| Sample | 10×Buffer | Enzyme 1 | Enzyme 2 | MilliQ | Total | Incubation | |
|---|---|---|---|---|---|---|---|
| S1 | 11µl | 5 | EcoRI 0.2 | SpeI 0.2 | 33.6 | 50 | At 37℃ for 2h |
| S2 | 11 | 5 | EcoRI 0.2 | SpeI 0.2 | 33.6 | 50 | |
| <partinfo>E0840</partinfo>(GFP) | 45 | 5 | EcoRI 0.2 | XbaI 0.2 | 0 | 50 |
After PCR purification, evaporated them and diluted 3ul.
7. Ligated over night.
| Sample | Vector | Insert | Ligation High | Total |
|---|---|---|---|---|
| S-GFP1 | <partinfo>E0840</partinfo> 0.5µl | S1 0.5 | 1 | 2 |
| S-GFP2 | <partinfo>E0840</partinfo> 0.5 | S2 0.5 | 1 | 2 |
Monday, July 26
By: Wataru, Tomonori, Makoto
1. Electrophoresis of PCR products
At the condition 4 (4.5µl MgSO4) and 6 (6µl MgSO4), S-R-Rz/Rz1 is amplified very much. So we decided to use them.
2. PCR purification
| Sample | Concentration (ng/µl) | New Name |
|---|---|---|
| 4 | 51.6 | |
| 5 | 59.3 | |
| 6 | 59.6 |
3. Transformation of iGEM Parts
| Name | Well | Sample (µl) | Competent Cell (µl) | Total (µl) | Plate | Incubation | Result |
|---|---|---|---|---|---|---|---|
| 1-12-M | 1 | 20 | 21 | LB (Amplicillin+) | At 37℃ 7/26 - 7/27 | × | |
| 2-17-F | 1 | 20 | 21 | LB (Kanamycin+) | × | ||
| 1-5-E | 1 | 20 | 21 | × |
 "
"