Team:HKU-Hong Kong/Project
From 2010.igem.org
(→The plasmid design) |
|||
| Line 63: | Line 63: | ||
==The plasmid design== | ==The plasmid design== | ||
| - | [[Image: | + | [[Image:gene E and lac operon.JPG|700px]] |
*placBAD: a promoter inducible by lactose or arabinose | *placBAD: a promoter inducible by lactose or arabinose | ||
*cI: the inhibitor of the pR promoter | *cI: the inhibitor of the pR promoter | ||
Revision as of 14:03, 13 October 2010

Several ideas on the subject of this year's investigation were come up during various brainstorming sessions. Our team picked out the idea of engineering a mechanism that can act as a safety net to prevent genetically modified bacteria from performing undesired tasks in wrong environments.
Bacteria are genetically engineered to perform various functions.Prospective functions include biodegradation of crude oil and killing of cancer cells. Yet, undesired tasks might be performed by the baterium itself as well. The idea of a "bio-safety" net would serve the purpose.
For example, after fulfillment of intended functions or when bacteria have escaped from the intented work zone.
Contents |
Overall project
ways to achieve this
- Insertion of killing genes
- Regulated by specific promoters
- Promoters respond to changes in the environment
Hence bacteria can and can only function well and survive in a specific environment
Project Details
Killing Mechanism
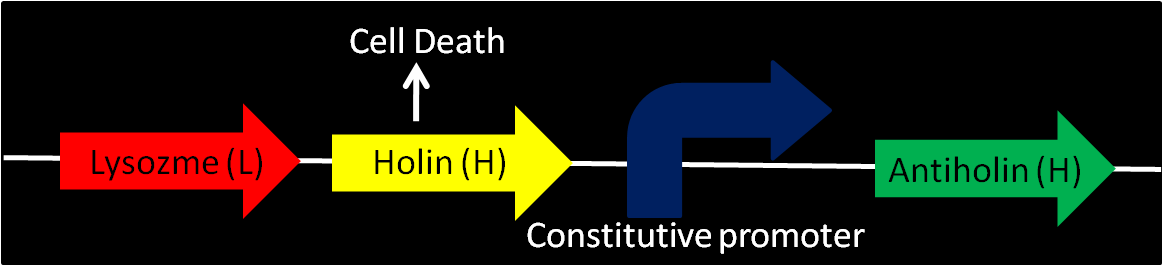
- Enterobacteriophage T4 Lysis Device
- We found the biobrick form Berkeley's team from iGEM 2008 a possible way to attain cell destruction.
Uses genes from enterobacteriaphage T4 (endolysin – lysozyme, holin and antiholin). Various promoters can be installed into this device to control lysis. Antiholin has its own constitutive promoter to prevent formation of holin multimers from basal expression. When device is off, higher expression of antiholin is needed for better stability of the device.
- Others
- Lytic enzyme systems from bacteriophage
- Lambda
- phiX174
- MS2
- Qβ
- T2
- Hybrid designer lytic enzyme
- Lytic enzyme systems from bacteriophage
Regulation
Placement
Changes in our plan
Difficulties
- Oil is a mixture of chemicals
- Does not induce promoters
- Difficult to target a specific component in oil
- Dfficult to implement the oil degrading system into E. Coli within the time period available to our project
The changes
After identification of the difficulties, we decided to change our plan and instead of focusing on oil, we will use lactose to imitate ‘oil’. This lactose analogue can also be substituted with different subtracts with appropriate modification on the biobricks in initiating the mechanism. Lactose is used as the analogue owing to various reasons:
- Lactose operon is widely studied
- No reporter needed
- Lac-Z assay
- Optimal difficulty and achievable for 2 months’ time
So, now our plan has changed from making an oil digesting bacterium to a lactose digesting bacterium. After the digestion is done, the bacteria will kill themselves.
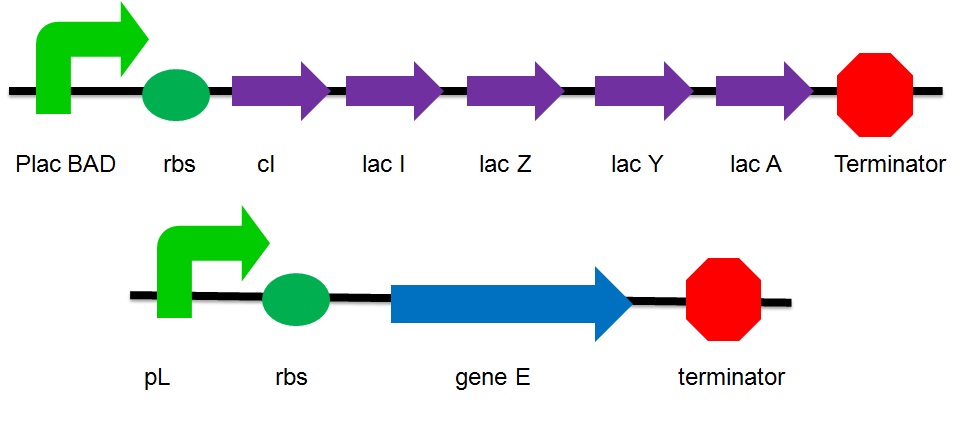
The plasmid design
- placBAD: a promoter inducible by lactose or arabinose
- cI: the inhibitor of the pR promoter
- Lac I: The inhibitor of the lac operon
- Lac Z: Beta-galactosidase
- Lac A : lactose acetylase
- Lac Y : lactose permease
- pL : a promoter that can be inhibited by cI
Scenarios
- In the presence of ‘oil’ (lactose), cI and the lac operon is expressed. ‘Oil’ (lactose) is digested
- When ‘oil’(lactose) is digested, cI is not produced. pL is no longer inhibited. The expression of the suicide genes will start leading to cell death.
- During the culture of the bacteria, arabinose is added to the LB agar, so that cI will be produced and the bacteria will not die.
Possible flaws
- Leaky expression
- The suicide genes won’t work
- The time it takes for cell death
Results
Possible further research
 "
"