Team:TU Delft/19 August 2010 content
From 2010.igem.org
(→Software development) |
(→Software development) |
||
| Line 9: | Line 9: | ||
The resulting list of interactions might be usable to 'debug' the resulting bacteria, i.a.w. suggest reasons why the system is not working like it should. Even though there would be no time to fix those problems, it could still help teams in following years that want to take another stab at solving the problem. | The resulting list of interactions might be usable to 'debug' the resulting bacteria, i.a.w. suggest reasons why the system is not working like it should. Even though there would be no time to fix those problems, it could still help teams in following years that want to take another stab at solving the problem. | ||
| - | Today the work on this application resulted in the first list of some potentially interacting proteins. A locally running version of the [http://string-db.org STRING protein database | + | Today the work on this application resulted in the first list of some potentially interacting proteins. A locally running version of the [http://string-db.org STRING protein database] (~150 gb PostgreSQL database) was used to look for interactions and homologs . Ideally however, this application would work using existing biological databases. |
I'm still looking for the right webservices to accomplish this, they might not exist. | I'm still looking for the right webservices to accomplish this, they might not exist. | ||
Revision as of 21:13, 22 August 2010
Software development
By Jelmer
Part of the dry lab work involves developing an application that can suggest possible interactions between the newly introduced proteins, and the existing E. coli proteome. This page will be updated with the application, documentation and results of applying it to our biobricks.
The basic idea is to look for interactions of the proteins within their own organism, and map those interactions by looking at homologs in E. coli:
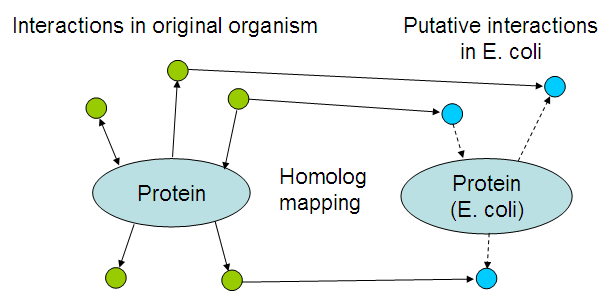
The resulting list of interactions might be usable to 'debug' the resulting bacteria, i.a.w. suggest reasons why the system is not working like it should. Even though there would be no time to fix those problems, it could still help teams in following years that want to take another stab at solving the problem.
Today the work on this application resulted in the first list of some potentially interacting proteins. A locally running version of the [http://string-db.org STRING protein database] (~150 gb PostgreSQL database) was used to look for interactions and homologs . Ideally however, this application would work using existing biological databases. I'm still looking for the right webservices to accomplish this, they might not exist.
 "
"