Team:Stockholm/21 August 2010
From 2010.igem.org
(→Gel verification) |
(→Assembly of CPP⋅protein⋅His constructs) |
||
| Line 26: | Line 26: | ||
*pSB1K3.His⋅yCCS: 1=A10, 2=A11, 3=A12 | *pSB1K3.His⋅yCCS: 1=A10, 2=A11, 3=A12 | ||
*Blank: A13 | *Blank: A13 | ||
| - | |||
| - | |||
| - | |||
| - | |||
Procedures as described in colony PCR protocol. | Procedures as described in colony PCR protocol. | ||
| Line 35: | Line 31: | ||
*Reverse primer: VR | *Reverse primer: VR | ||
*Elongation time: 2:00 | *Elongation time: 2:00 | ||
| + | |||
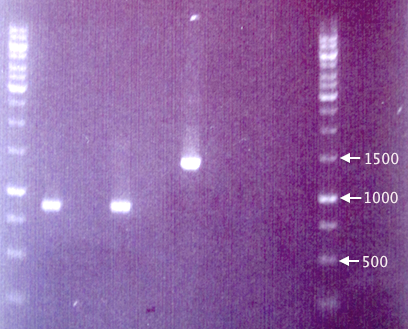
| + | =====Gel verification===== | ||
| + | [[image:ColPCR_pEX.RFP_21aug.png|200px|thumb|right|'''Gel verification of pEX.RFP constructs and pEX vector.'''<br />'''Loading:''' λ, A1, A8, A2, A3, A4, A5, A6, A7, λ<br />3 μl λ, 4 μl sample. λ = O'GeneRuler 1 kb DNA ladder.]] | ||
| + | 1 % agarose, 90 V, 1 h. | ||
====Transfer of RFP to pEX==== | ====Transfer of RFP to pEX==== | ||
Revision as of 17:08, 22 August 2010
Contents |
Hassan
found some extra articles for gene-regulatory networks: [http://www.ncbi.nlm.nih.gov/pubmed/17504165] and [http://www.biomedcentral.com/1471-2105/8/S6/S9]
Andreas
Assembly of CPP⋅protein⋅His constructs
Assembly of His⋅SOD/yCCS and SOD/yCCS⋅His into pSB1K3
Step I in cloning strategy (19/8)
Transformation results
- pSB1K3.SOD⋅His
- pSB1K3.yCCS⋅His
- pSB1K3.His⋅SOD
- pSB1K3.His⋅yCCS
Good yield on all plates.
Colony PCR
Three white colonies picked from each plate for verification.
- pSB1K3.SOD⋅His: 1=A1, 2=A2, 3=A3
- pSB1K3.yCCS⋅His: 1=A4, 2=A5, 3=A6
- pSB1K3.His⋅SOD: 1=A7, 2=A8, 3=A9
- pSB1K3.His⋅yCCS: 1=A10, 2=A11, 3=A12
- Blank: A13
Procedures as described in colony PCR protocol.
- Forward primer: VF2
- Reverse primer: VR
- Elongation time: 2:00
Gel verification
1 % agarose, 90 V, 1 h.
Transfer of RFP to pEX
Continued from 19/8
Colony PCR
Since the gel results from 20/8 only verified pMA.His, a new colony PCR was run to verify pEX and pEX.RFP. Two new clones (5 & 6) of pEX.RFP were picked and run with clones 2 and 3 from 20/8. To test whether the red colonies could possibly be from pSB1K3 (i.e. if the selection didn't work properly), pEX.RFP 2 was also amplified with pSB primer VF2 and VR.
Also, pEX plasmid was amplified in parallel with pEX plasmid to compare the insert sizes.
- J1: pEX.RFP 2
- J8: pEX.RFP 3
- J2: pEX.RFP 5
- J3: pEX.RFP 6
- J4: pEX clone
- J5: pEX plasmid
- J6: pEX.RFP 2 (w/ pSB primers)
- J7: Blank
Procedures as described in colony PCR protocol:
- Forward primer: pEXf or pSB VF2
- Reverse primer: pEXr or pSB VR
- Elongation: 2:00
Gel verification
To be added
Colony re-streak
In order to verify that our red colonies are indeed carrying pEX and not pSB1K3 (i.e. the Amp selection has failed), resulting in false-positives for transfer of RFP from pSB1K3 to pEX, clones 2, 3, 5 and 6 were streaked on quarters of Amp 100 and Km 50 plates. Plates incubated 48 h in 37 °C.
If RFP has been successfully transfered to pEX, the clones should only grow on Amp 100 plates.
 "
"