Filamentous Cells
From 2010.igem.org
(Difference between revisions)
RachelBoyd (Talk | contribs) |
RachelBoyd (Talk | contribs) |
||
| Line 8: | Line 8: | ||
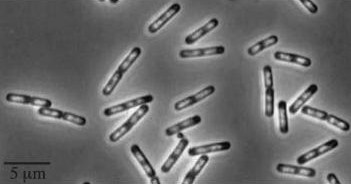
| Wild type Bacillus subtillis [[Image:Wild type Bacillus subtillis.jpg]] | | Wild type Bacillus subtillis [[Image:Wild type Bacillus subtillis.jpg]] | ||
|- | |- | ||
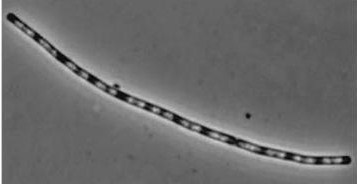
| - | |dinR KO [[Image:dinR KO]] | + | |dinR KO [[Image:dinR KO.jpg]] |
|- | |- | ||
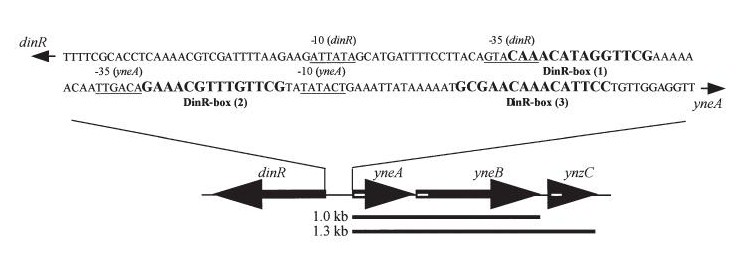
| - | |dinR KO mutant over expressed the divergent (opposite direction) transcript for YneA, YneB and YnzC. These genes form the SOS regulon (recA independent SOS response). [[Image:Coding region]] | + | |dinR KO mutant over expressed the divergent (opposite direction) transcript for YneA, YneB and YnzC. These genes form the SOS regulon (recA independent SOS response). [[Image:Coding region.jpg]] |
|- | |- | ||
|YneA suppressed in wt without SOS induction | |YneA suppressed in wt without SOS induction | ||
| Line 18: | Line 18: | ||
|Disruption of YneA in SOS response leads to reduced elongation. Altering YneB and YnzC expression does not affect cell morphology. | |Disruption of YneA in SOS response leads to reduced elongation. Altering YneB and YnzC expression does not affect cell morphology. | ||
|- | |- | ||
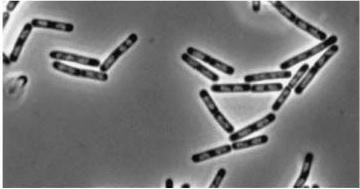
| - | |Double mutant (dinR/YneA) [[Image:Double Mutant]] | + | |Double mutant (dinR/YneA) [[Image:Double Mutant.jpg]] |
|- | |- | ||
|YneA protein required to suppress cell division. Not chromosome replication or segregation. | |YneA protein required to suppress cell division. Not chromosome replication or segregation. | ||
Revision as of 12:25, 29 May 2010
Filamentous Cells
 "
"