Team:TU Delft/23 July 2010 content
From 2010.igem.org
Ravandervalk (Talk | contribs) (→Salt tolerance) |
Ravandervalk (Talk | contribs) (→Salt tolerance) |
||
| Line 191: | Line 191: | ||
The paltes transformed on wednesday had colonies but they grew very slowly. Thus they were grown up for another night and were Sc PCR'd today. | The paltes transformed on wednesday had colonies but they grew very slowly. Thus they were grown up for another night and were Sc PCR'd today. | ||
| + | [[Image:Tudelft_ScPCR_bbc1-B0015.jpg|400px]] | ||
| + | |||
| + | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
| + | |'''slot #''' | ||
| + | |'''Contents''' | ||
| + | |'''Result''' | ||
| + | |- | ||
| + | |1 | ||
| + | |NC (RFP from resealing plasmids) | ||
| + | | | ||
| + | |- | ||
| + | |2 | ||
| + | |Ez Load | ||
| + | | | ||
| + | |- | ||
| + | |3 | ||
| + | |SC Sample 1 | ||
| + | |Possibly positive | ||
| + | |- | ||
| + | |4 | ||
| + | |Sc Sample 2 | ||
| + | |Negative | ||
| + | |- | ||
| + | |5 | ||
| + | |Sc Sample 3 | ||
| + | |Negative | ||
| + | |- | ||
| + | |6 | ||
| + | |Sc Sample 4 | ||
| + | |Possibly positive | ||
| + | |- | ||
| + | |7 | ||
| + | |SC Sample 5 | ||
| + | |Possibly positive | ||
| + | |- | ||
| + | |8 | ||
| + | |Smartladder | ||
| + | |} | ||
| + | |||
| + | |||
| + | Here you can see that 3 samples are possibly positive, these will be sequenced and used for further steps in this process. | ||
Latest revision as of 11:18, 10 August 2010
Contents |
Surprise
We are very grateful for the help of our superior, Esengül in the lab. To thank her, we organized a surprise for her… In the afternoon a special delivery for Esengül arrived. Inside was a self-baked cake from a secret admirer. What a surprise this was! Half a day later, she found out that we were behind it and we all enjoyed eating the cake.
Lab work
Alkane Degradation
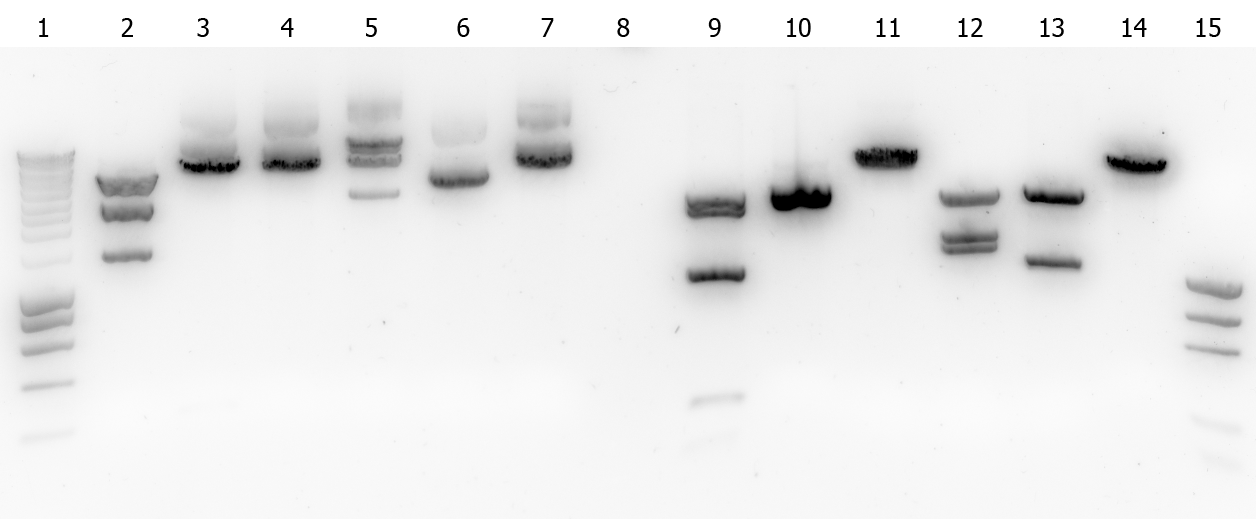
With a number of yesterday's colonies a digestion was done to check if they contain the desired insert. The table below describes the digestion procedure, as well as the gel lane description. (Digestion was done in 10 μL end-volume)
| # | Sample | Enzyme 1 | Enzyme 2 | Buffer | BSA |
| 1 | 007T (2) | 0.5 μL EcoRI | 0.5 μL PstI | 3 (Biolabs) | ✗ |
| 2 | 008T (4) | 0.5 μL EcoRI | 0.5 μL PstI | 3 (Biolabs) | ✗ |
| 3 | 009T (6) | 0.5 μL EcoRI | 0.5 μL PstI | 3 (Biolabs) | ✗ |
| 4 | 009T (7) | 0.5 μL EcoRI | 0.5 μL PstI | 3 (Biolabs) | ✗ |
| 5 | 010T (9) | 0.5 μL EcoRI | 0.5 μL PstI | 3 (Biolabs) | ✗ |
| 6 | 017T (14) | 0.5 μL EcoRI | 0.5 μL PstI | 3 (Biolabs) | ✗ |
| 7 | 007T (2) | 1 μL PstI | 1 μL PvuII | H (Boehringer) | ✗ |
| 8 | 008T (4) | 1 μL AatII | 0.5 μL KpnI | A (Boehringer) | ✓ |
| 9 | 009T (6) | 0.5 μL KpnI | 1 μL NruI | B (Boehringer) | ✓ |
| 10 | 009T (7) | 0.5 μL KpnI | 1 μL NruI | B (Boehringer) | ✓ |
| 11 | 010T (9) | 1 μL EarI | 0.5 μL PstI | 1 (Biolabs) | ✗ |
| 12 | 017T (14) | 0.5 μL AdhI | 0.5 μL SpeI | 4 (Biolabs) | ✗ |
Lane description:
| # | Description | Expected Length (bp) | Status |
| M1 | SmartLadder | n/a | n/a |
| 1 | 007T (2) + EcoRI + PstI | 1341, 2455 | |
| 2 | 008T (4) + EcoRI + PstI | 276, 2455 | |
| 3 | 009T (6) + EcoRI + PstI | 285, 2455 | |
| 4 | 009T (7) + EcoRI + PstI | 285, 2455 | |
| 5 | 010T (9) + EcoRI + PstI | 1382, 2455 | |
| 6 | 017T (14) + EcoRI + PstI | 855, 2455 | |
| 7 | Empty | ||
| 8 | 007T (2) + PstI + PvuII | 2529,983,251 | |
| 9 | 008T (4) + AatII + KpnI | 271, 2427 | |
| 10 | 009T (6) + KpnI + NruI | 598, 2109 | |
| 11 | 009T (7) + KpnI + NruI | 598, 2109 | |
| 12 | 010T (9) + EarI + PstI | 1075, 2729 | |
| 13 | 017T (14) + AdhI + SpeI | 428, 2849 | |
| M2 | EZ load (ladder) | n/a | n/a |
It seems (from the gel) that BioBrick 007T is the correct one, and the others don't contain the desired insert.
Salt tolerance
The paltes transformed on wednesday had colonies but they grew very slowly. Thus they were grown up for another night and were Sc PCR'd today.
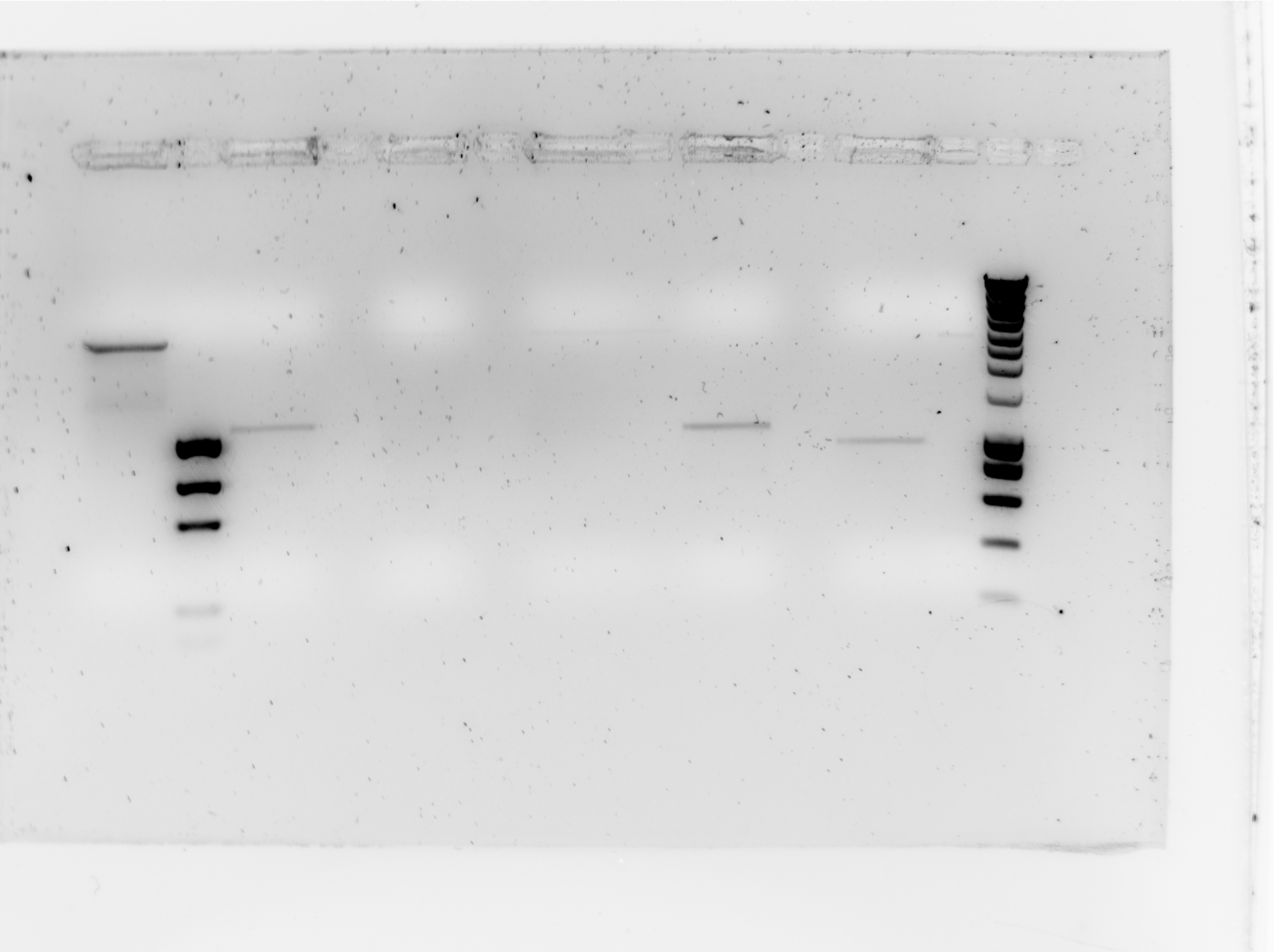
| slot # | Contents | Result |
| 1 | NC (RFP from resealing plasmids) | |
| 2 | Ez Load | |
| 3 | SC Sample 1 | Possibly positive |
| 4 | Sc Sample 2 | Negative |
| 5 | Sc Sample 3 | Negative |
| 6 | Sc Sample 4 | Possibly positive |
| 7 | SC Sample 5 | Possibly positive |
| 8 | Smartladder |
Here you can see that 3 samples are possibly positive, these will be sequenced and used for further steps in this process.
 "
"